Consider the following, simple model:
import pymc3 as pm
import numpy as np
with pm.Model() as model:
pm.Binomial('scalar_binomial', n=1, p=0.25)
pm.Binomial('arrayed_binomial', n=1, p=0.25, shape=10)
pm.Binomial(
'arrayed_binomial_alt', n=np.ones(10),
p=np.full(10, 0.25), shape=10)
trace = pm.sample(draws=600, chains=2, tune=500)
summary = pm.summary(
trace,
var_names=[
'scalar_binomial', 'arrayed_binomial',
'arrayed_binomial_alt'])
summary
The three RVs are identical binomials, except that the first scalar_binomial
is a scalar, the second arrayed_binomial has shape 10 with scalar arguments,
and the third arrayed_binomial_alt has shape 10 with (identical) arrayed
arguments. Note also that nothing in this model is constrained by observations.
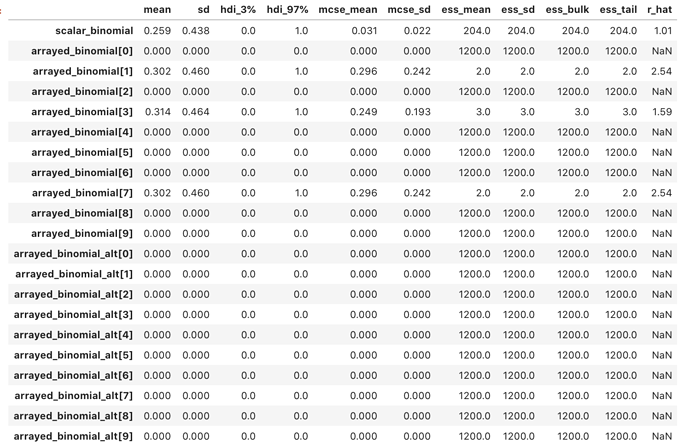
The result of sampling is peculiar:
scalar_binomial is as expected. arrayed_binomial sees 7 of the 10 array
elements with no binomial samples, and means of 0.0. arrayed_binomial_alt
sees all 10 array elements with no binomial samples, and means of 0.0.
Is this a bug with pm.Binomial()? But how could a bug on something this basic escape notice? So perhaps I am misunderstanding.
The same model structure with normals instead of binomials produces expected
results:
import pymc3 as pm
import numpy as np
with pm.Model() as model:
pm.Normal('scalar_normal', mu=1, sigma=0.25)
pm.Normal('arrayed_normal', mu=1, sigma=0.25, shape=10)
pm.Normal(
'arrayed_normal_alt', mu=np.ones(10),
sigma=np.full(10, 0.25), shape=10)
trace = pm.sample(
draws=600, chains=2, tune=500,
step=pm.Metropolis()) # Metropolis so the sampling is identical to above
summary = pm.summary(
trace,
var_names=[
'scalar_normal', 'arrayed_normal', 'arrayed_normal_alt'])
summary
Results, as expected:
Configuration:
PyMC3 v3.9.3
Numpy v1.19.4
Theano v1.0.5