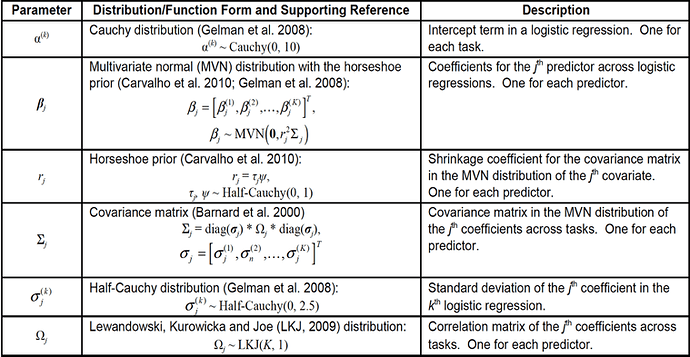
I was trying to build a hierarchical bayesian logistic classification model. I regard 5 binary classification tasks as different but related tasks. The 5 tasks have the same 24 predicators.The regression coefficients for the same predictor across all tasks are multivariate normally (MVN) distributed, with zero means as well as a scaled covariance matrix.

I failed to find a simple way to set the dimensions of the LKJ distribution as i need so I set them separately. Here is the code:
with pm.Model() as model:
# priors on shrikage scalar
psi = pm.HalfCauchy('psi', beta=1, shape=1)
tau = pm.HalfCauchy('tau', beta=1, shape=num_features)
r_shrinkage = pm.Deterministic('r', psi*tau) # shrinkage scalar for each predictor
r_square = pm.Deterministic('r_square', np.multiply(r_shrinkage,r_shrinkage))
#priors on standard deviation
sigma_1 = pm.HalfCauchy('sigma_1', beta=2.5, shape=num_tasks)
sigma_2 = pm.HalfCauchy('sigma_2', beta=2.5, shape=num_tasks)
sigma_3 = pm.HalfCauchy('sigma_3', beta=2.5, shape=num_tasks)
sigma_4 = pm.HalfCauchy('sigma_4', beta=2.5, shape=num_tasks)
sigma_5 = pm.HalfCauchy('sigma_5', beta=2.5, shape=num_tasks)
sigma_6 = pm.HalfCauchy('sigma_6', beta=2.5, shape=num_tasks)
sigma_7 = pm.HalfCauchy('sigma_7', beta=2.5, shape=num_tasks)
sigma_8 = pm.HalfCauchy('sigma_8', beta=2.5, shape=num_tasks)
sigma_9 = pm.HalfCauchy('sigma_9', beta=2.5, shape=num_tasks)
sigma_10 = pm.HalfCauchy('sigma_10', beta=2.5, shape=num_tasks)
sigma_11 = pm.HalfCauchy('sigma_11', beta=2.5, shape=num_tasks)
sigma_12 = pm.HalfCauchy('sigma_12', beta=2.5, shape=num_tasks)
sigma_13 = pm.HalfCauchy('sigma_13', beta=2.5, shape=num_tasks)
sigma_14 = pm.HalfCauchy('sigma_14', beta=2.5, shape=num_tasks)
sigma_15 = pm.HalfCauchy('sigma_15', beta=2.5, shape=num_tasks)
sigma_16 = pm.HalfCauchy('sigma_16', beta=2.5, shape=num_tasks)
sigma_17 = pm.HalfCauchy('sigma_17', beta=2.5, shape=num_tasks)
sigma_18 = pm.HalfCauchy('sigma_18', beta=2.5, shape=num_tasks)
sigma_19 = pm.HalfCauchy('sigma_19', beta=2.5, shape=num_tasks)
sigma_20 = pm.HalfCauchy('sigma_20', beta=2.5, shape=num_tasks)
sigma_21 = pm.HalfCauchy('sigma_21', beta=2.5, shape=num_tasks)
sigma_22 = pm.HalfCauchy('sigma_22', beta=2.5, shape=num_tasks)
sigma_23 = pm.HalfCauchy('sigma_23', beta=2.5, shape=num_tasks)
sigma_24 = pm.HalfCauchy('sigma_24', beta=2.5, shape=num_tasks)
#LKJ prior for correlation matrix as upper triangular vector
C_triu_1 = pm.LKJCorr('C_triu_1', eta=1, n=num_tasks)
C_triu_2 = pm.LKJCorr('C_triu_2', eta=1, n=num_tasks)
C_triu_3 = pm.LKJCorr('C_triu_3', eta=1, n=num_tasks)
C_triu_4 = pm.LKJCorr('C_triu_4', eta=1, n=num_tasks)
C_triu_5 = pm.LKJCorr('C_triu_5', eta=1, n=num_tasks)
C_triu_6 = pm.LKJCorr('C_triu_6', eta=1, n=num_tasks)
C_triu_7 = pm.LKJCorr('C_triu_7', eta=1, n=num_tasks)
C_triu_8 = pm.LKJCorr('C_triu_8', eta=1, n=num_tasks)
C_triu_9 = pm.LKJCorr('C_triu_9', eta=1, n=num_tasks)
C_triu_10 = pm.LKJCorr('C_triu_10', eta=1, n=num_tasks)
C_triu_11 = pm.LKJCorr('C_triu_11', eta=1, n=num_tasks)
C_triu_12 = pm.LKJCorr('C_triu_12', eta=1, n=num_tasks)
C_triu_13 = pm.LKJCorr('C_triu_13', eta=1, n=num_tasks)
C_triu_14 = pm.LKJCorr('C_triu_14', eta=1, n=num_tasks)
C_triu_15 = pm.LKJCorr('C_triu_15', eta=1, n=num_tasks)
C_triu_16 = pm.LKJCorr('C_triu_16', eta=1, n=num_tasks)
C_triu_17 = pm.LKJCorr('C_triu_17', eta=1, n=num_tasks)
C_triu_18 = pm.LKJCorr('C_triu_18', eta=1, n=num_tasks)
C_triu_19 = pm.LKJCorr('C_triu_19', eta=1, n=num_tasks)
C_triu_20 = pm.LKJCorr('C_triu_20', eta=1, n=num_tasks)
C_triu_21 = pm.LKJCorr('C_triu_21', eta=1, n=num_tasks)
C_triu_22 = pm.LKJCorr('C_triu_22', eta=1, n=num_tasks)
C_triu_23 = pm.LKJCorr('C_triu_23', eta=1, n=num_tasks)
C_triu_24 = pm.LKJCorr('C_triu_24', eta=1, n=num_tasks)
#convert to matrix form
C_1 = T.fill_diagonal(C_triu_1[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_2 = T.fill_diagonal(C_triu_2[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_3 = T.fill_diagonal(C_triu_3[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_4 = T.fill_diagonal(C_triu_4[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_5 = T.fill_diagonal(C_triu_5[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_6 = T.fill_diagonal(C_triu_6[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_7 = T.fill_diagonal(C_triu_7[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_8 = T.fill_diagonal(C_triu_8[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_9 = T.fill_diagonal(C_triu_9[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_10 = T.fill_diagonal(C_triu_10[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_11 = T.fill_diagonal(C_triu_11[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_12 = T.fill_diagonal(C_triu_12[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_13 = T.fill_diagonal(C_triu_13[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_14 = T.fill_diagonal(C_triu_14[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_15 = T.fill_diagonal(C_triu_15[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_16 = T.fill_diagonal(C_triu_16[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_17 = T.fill_diagonal(C_triu_17[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_18 = T.fill_diagonal(C_triu_18[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_19 = T.fill_diagonal(C_triu_19[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_20 = T.fill_diagonal(C_triu_20[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_21 = T.fill_diagonal(C_triu_21[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_22 = T.fill_diagonal(C_triu_22[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_23 = T.fill_diagonal(C_triu_23[np.zeros((num_tasks,num_tasks), 'int')], 1)
C_24 = T.fill_diagonal(C_triu_24[np.zeros((num_tasks,num_tasks), 'int')], 1)
sigma_diag_1 = T.nlinalg.diag(sigma_1)
sigma_diag_2 = T.nlinalg.diag(sigma_2)
sigma_diag_3 = T.nlinalg.diag(sigma_3)
sigma_diag_4 = T.nlinalg.diag(sigma_4)
sigma_diag_5 = T.nlinalg.diag(sigma_5)
sigma_diag_6 = T.nlinalg.diag(sigma_6)
sigma_diag_7 = T.nlinalg.diag(sigma_7)
sigma_diag_8 = T.nlinalg.diag(sigma_8)
sigma_diag_9 = T.nlinalg.diag(sigma_9)
sigma_diag_10 = T.nlinalg.diag(sigma_10)
sigma_diag_11 = T.nlinalg.diag(sigma_11)
sigma_diag_12 = T.nlinalg.diag(sigma_12)
sigma_diag_13 = T.nlinalg.diag(sigma_13)
sigma_diag_14 = T.nlinalg.diag(sigma_14)
sigma_diag_15 = T.nlinalg.diag(sigma_15)
sigma_diag_16 = T.nlinalg.diag(sigma_16)
sigma_diag_17 = T.nlinalg.diag(sigma_17)
sigma_diag_18 = T.nlinalg.diag(sigma_18)
sigma_diag_19 = T.nlinalg.diag(sigma_19)
sigma_diag_20 = T.nlinalg.diag(sigma_20)
sigma_diag_21 = T.nlinalg.diag(sigma_21)
sigma_diag_22 = T.nlinalg.diag(sigma_22)
sigma_diag_23 = T.nlinalg.diag(sigma_23)
sigma_diag_24 = T.nlinalg.diag(sigma_24)
#induced covariance matrix
cov_1 = pm.Deterministic('cov_1', T.nlinalg.matrix_dot(sigma_diag_1, C_1, sigma_diag_1))
cov_2 = pm.Deterministic('cov_2', T.nlinalg.matrix_dot(sigma_diag_2, C_2, sigma_diag_2))
cov_3 = pm.Deterministic('cov_3', T.nlinalg.matrix_dot(sigma_diag_3, C_3, sigma_diag_3))
cov_4 = pm.Deterministic('cov_4', T.nlinalg.matrix_dot(sigma_diag_4, C_4, sigma_diag_4))
cov_5 = pm.Deterministic('cov_5', T.nlinalg.matrix_dot(sigma_diag_5, C_5, sigma_diag_5))
cov_6 = pm.Deterministic('cov_6', T.nlinalg.matrix_dot(sigma_diag_6, C_6, sigma_diag_6))
cov_7 = pm.Deterministic('cov_7', T.nlinalg.matrix_dot(sigma_diag_7, C_7, sigma_diag_7))
cov_8 = pm.Deterministic('cov_8', T.nlinalg.matrix_dot(sigma_diag_8, C_8, sigma_diag_8))
cov_9 = pm.Deterministic('cov_9', T.nlinalg.matrix_dot(sigma_diag_9, C_9, sigma_diag_9))
cov_10 = pm.Deterministic('cov_10', T.nlinalg.matrix_dot(sigma_diag_10, C_10, sigma_diag_10))
cov_11 = pm.Deterministic('cov_11', T.nlinalg.matrix_dot(sigma_diag_11, C_11, sigma_diag_11))
cov_12 = pm.Deterministic('cov_12', T.nlinalg.matrix_dot(sigma_diag_12, C_12, sigma_diag_12))
cov_13 = pm.Deterministic('cov_13', T.nlinalg.matrix_dot(sigma_diag_13, C_13, sigma_diag_13))
cov_14 = pm.Deterministic('cov_14', T.nlinalg.matrix_dot(sigma_diag_14, C_14, sigma_diag_14))
cov_15 = pm.Deterministic('cov_15', T.nlinalg.matrix_dot(sigma_diag_15, C_15, sigma_diag_15))
cov_16 = pm.Deterministic('cov_16', T.nlinalg.matrix_dot(sigma_diag_16, C_16, sigma_diag_16))
cov_17 = pm.Deterministic('cov_17', T.nlinalg.matrix_dot(sigma_diag_17, C_17, sigma_diag_17))
cov_18 = pm.Deterministic('cov_18', T.nlinalg.matrix_dot(sigma_diag_18, C_18, sigma_diag_18))
cov_19 = pm.Deterministic('cov_19', T.nlinalg.matrix_dot(sigma_diag_19, C_19, sigma_diag_19))
cov_20 = pm.Deterministic('cov_20', T.nlinalg.matrix_dot(sigma_diag_20, C_20, sigma_diag_20))
cov_21 = pm.Deterministic('cov_21', T.nlinalg.matrix_dot(sigma_diag_21, C_21, sigma_diag_21))
cov_22 = pm.Deterministic('cov_22', T.nlinalg.matrix_dot(sigma_diag_22, C_22, sigma_diag_22))
cov_23 = pm.Deterministic('cov_23', T.nlinalg.matrix_dot(sigma_diag_23, C_23, sigma_diag_23))
cov_24 = pm.Deterministic('cov_24', T.nlinalg.matrix_dot(sigma_diag_24, C_24, sigma_diag_24))
# intercept of logit model ----alpha
alpha = pm.Cauchy('alpha', alpha=0, beta=10, shape=num_tasks)
# params of logit model ----beta
beta_1 = pm.MvNormal('beta_1', mu=np.zeros(num_tasks), chol=T.dot(r_square[0],cov_1),shape=num_tasks)
beta_2 = pm.MvNormal('beta_2', mu=np.zeros(num_tasks), chol=T.dot(r_square[1],cov_2),shape=num_tasks)
beta_3 = pm.MvNormal('beta_3', mu=np.zeros(num_tasks), chol=T.dot(r_square[2],cov_3),shape=num_tasks)
beta_4 = pm.MvNormal('beta_4', mu=np.zeros(num_tasks), chol=T.dot(r_square[3],cov_4),shape=num_tasks)
beta_5 = pm.MvNormal('beta_5', mu=np.zeros(num_tasks), chol=T.dot(r_square[4],cov_5),shape=num_tasks)
beta_6 = pm.MvNormal('beta_6', mu=np.zeros(num_tasks), chol=T.dot(r_square[5],cov_6),shape=num_tasks)
beta_7 = pm.MvNormal('beta_7', mu=np.zeros(num_tasks), chol=T.dot(r_square[6],cov_7),shape=num_tasks)
beta_8 = pm.MvNormal('beta_8', mu=np.zeros(num_tasks), chol=T.dot(r_square[7],cov_8),shape=num_tasks)
beta_9 = pm.MvNormal('beta_9', mu=np.zeros(num_tasks), chol=T.dot(r_square[8],cov_9),shape=num_tasks)
beta_10 = pm.MvNormal('beta_10', mu=np.zeros(num_tasks), chol=T.dot(r_square[9],cov_10),shape=num_tasks)
beta_11 = pm.MvNormal('beta_11', mu=np.zeros(num_tasks), chol=T.dot(r_square[10],cov_11),shape=num_tasks)
beta_12 = pm.MvNormal('beta_12', mu=np.zeros(num_tasks), chol=T.dot(r_square[11],cov_12),shape=num_tasks)
beta_13 = pm.MvNormal('beta_13', mu=np.zeros(num_tasks), chol=T.dot(r_square[12],cov_13),shape=num_tasks)
beta_14 = pm.MvNormal('beta_14', mu=np.zeros(num_tasks), chol=T.dot(r_square[13],cov_14),shape=num_tasks)
beta_15 = pm.MvNormal('beta_15', mu=np.zeros(num_tasks), chol=T.dot(r_square[14],cov_15),shape=num_tasks)
beta_16 = pm.MvNormal('beta_16', mu=np.zeros(num_tasks), chol=T.dot(r_square[15],cov_16),shape=num_tasks)
beta_17 = pm.MvNormal('beta_17', mu=np.zeros(num_tasks), chol=T.dot(r_square[16],cov_17),shape=num_tasks)
beta_18 = pm.MvNormal('beta_18', mu=np.zeros(num_tasks), chol=T.dot(r_square[17],cov_18),shape=num_tasks)
beta_19 = pm.MvNormal('beta_19', mu=np.zeros(num_tasks), chol=T.dot(r_square[18],cov_19),shape=num_tasks)
beta_20 = pm.MvNormal('beta_20', mu=np.zeros(num_tasks), chol=T.dot(r_square[19],cov_20),shape=num_tasks)
beta_21 = pm.MvNormal('beta_21', mu=np.zeros(num_tasks), chol=T.dot(r_square[20],cov_21),shape=num_tasks)
beta_22 = pm.MvNormal('beta_22', mu=np.zeros(num_tasks), chol=T.dot(r_square[21],cov_22),shape=num_tasks)
beta_23 = pm.MvNormal('beta_23', mu=np.zeros(num_tasks), chol=T.dot(r_square[22],cov_23),shape=num_tasks)
beta_24 = pm.MvNormal('beta_24', mu=np.zeros(num_tasks), chol=T.dot(r_square[23],cov_24),shape=num_tasks)
# observed data
likelihood = pm.math.invlogit(alpha[industry_idx] + X_shared[:,0]*beta_1[industry_idx]+X_shared[:,1]*beta_2[industry_idx]+X_shared[:,2]*beta_3[industry_idx]+X_shared[:,3]*beta_4[industry_idx]+X_shared[:,4]*beta_5[industry_idx]+X_shared[:,5]*beta_6[industry_idx]+X_shared[:,6]*beta_7[industry_idx]+X_shared[:,7]*beta_8[industry_idx]+X_shared[:,8]*beta_9[industry_idx]+X_shared[:,9]*beta_10[industry_idx]+X_shared[:,10]*beta_11[industry_idx]+X_shared[:,11]*beta_12[industry_idx]+X_shared[:,12]*beta_13[industry_idx]+X_shared[:,13]*beta_14[industry_idx]+X_shared[:,14]*beta_15[industry_idx]+X_shared[:,15]*beta_16[industry_idx]+X_shared[:,16]*beta_17[industry_idx]+X_shared[:,17]*beta_18[industry_idx]+X_shared[:,18]*beta_19[industry_idx]+X_shared[:,19]*beta_20[industry_idx]+X_shared[:,20]*beta_21[industry_idx]+X_shared[:,21]*beta_22[industry_idx]+X_shared[:,22]*beta_23[industry_idx]+X_shared[:,23]*beta_24[industry_idx]) # calculate fraud probability
y_fraud = pm.Bernoulli('fraud', p=likelihood, observed=y_train)
When I run the code above with NUTs sampler, it tirggers the ValueError:Bad initial energy: inf. The model might be misspecified.
Thanks for your help!