I have a NN implemented using Theano and PyMC3. The network architecture is specified as such:
with pm.Model() as nn_model:
n_hidden = new_df.features().shape[1]
w1 = pm.Normal('w1', mu=0, sd=1, shape=(new_df.features().shape[1], n_hidden))
w2 = pm.Normal('w2', mu=0, sd=1, shape=(n_hidden, 1))
b1 = pm.Normal('b1', mu=0, sd=1, shape=(n_hidden,))
b2 = pm.Normal('b2', mu=0, sd=1, shape=(1,))
a1 = pm.Deterministic('a1', tt.nnet.relu(tt.dot(X, w1) + b1))
a2 = pm.Deterministic('a2', tt.nnet.sigmoid(tt.dot(a1, w2) + b2))
out = pm.Bernoulli('likelihood', p=a2, observed=Y)
Vanilla feed-forward neural network, nothing fancy.
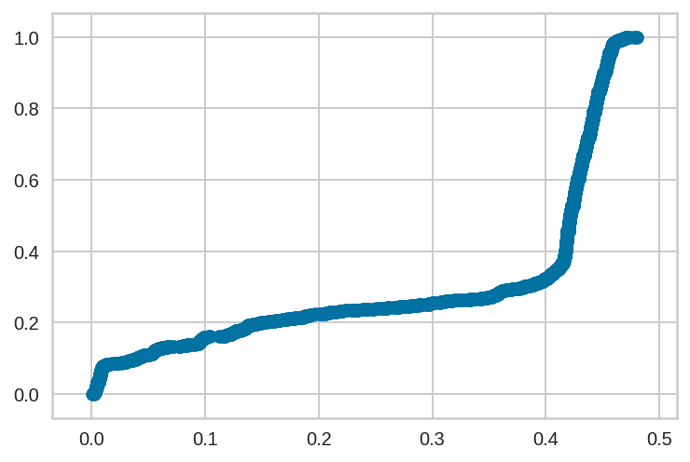
One thing I did notice that was weird after fitting with ADVI, the prediction probability range was between 0 and 0.5, rather than 0 and 1. See ECDF below.
The distribution of true/false does mimic the population distribution of true/false, just that the predictions are squished between 0 and 0.5, rather than 0 and 1.
I found that kind of weird; is there a way to diagnose how this is occurring?
 . Maybe I’ll change my floatX to see if that changes things? I’m using theano 0.9.0 right now.
. Maybe I’ll change my floatX to see if that changes things? I’m using theano 0.9.0 right now.