Hello,
I am trying to deal with my missing data of the target values by using mask, and I want to calculate the likelihood just at the points where the target values are missing, how should I do?
https://github.com/pymc-devs/pymc3/blob/master/pymc3/examples/lasso_missing.py
My model looks like this.
ann_input = theano.shared(X_train)
ann_output = theano.shared(Y_train)
with pm.Model() as neural_network:
weights_in_1 = pm.Normal('w_in_1', 0, sd = 1, shape=(5,3), testval=init_1)
weights_1_out = pm.Normal('w_1_out', 0, sd = 1, shape=(3,), testval=init_out)
hidden1_bias = pm.Normal( 'hidden1_bias', sd=1, shape=n_hidden1)
hidden_out_bias = pm.Normal('hidden_out_bias', mu = 3, sd=1)
act_1 = pm.math.tanh( pm.math.dot(ann_input, weights_in_1 )+ hidden1_bias[None, :])
regression = T.dot(act_1, weights_1_out) + hidden_out_bias
pm.Normal('out', mu =regression, sd=np.sqrt( 0.9 ), observed = ann_output)
train_trace = pm.sample( )
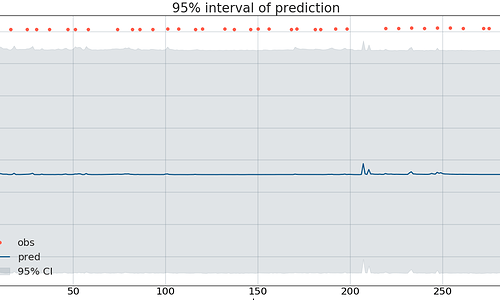
In Likelihood function ann_output has some values masked, and part of it looks like this
Based on the example from disaster case study, I changed the value to -999 for mask, then I got the result
So I wonder if the data was really masked, I changed to 999, and mask the value 999, then I got the result.