I have a change point model in Stan
data {
int<lower=1> N;
vector[N] x;
vector[N] y;
}
parameters {
real tau;
real mu1;
real mu2;
real gamma1;
real gamma2;
real<lower=0> sigma1;
real<lower=0> sigma2;
}
model {
real mu;
real gamma;
real sigma;
mu1 ~ normal(0, 10);
mu2 ~ normal(0, 10);
gamma1 ~ normal(0, 10);
gamma2 ~ normal(0, 10);
sigma1 ~ normal(0, 10);
sigma2 ~ normal(0, 10);
tau ~ uniform(0,N+1);
for (i in 1:N) {
mu = i < tau ? mu1 : mu2;
gamma = i < tau ? gamma1 : gamma2;
sigma = i < tau ? sigma1 : sigma2;
y[i] ~ normal(mu * x[i] + gamma, sigma);
}
}
which with some generated data in R
x <- c(1:100)
set.seed(42)
z1 <- rnorm(50,0.0,2.1)
z2 <- rnorm(50,0.0,2.2)
mu1 <- 1.0
mu2 <- 2.0
gamma1 <- 10.0
gamma2 <- -40.0
y1 <- mu1 * x[1:50] + gamma1 + z1
y2 <- mu2 * x[51:100] + gamma2 + z2
y <- c(y1,y2)
gives a good fit
Inference for Stan model: lr-changepoint-cont.
4 chains, each with iter=10000; warmup=1000; thin=1;
post-warmup draws per chain=9000, total post-warmup draws=36000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
tau 47.93 0.05 2.50 43.42 46.18 47.96 49.63 52.71 2414 1
mu1 0.96 0.00 0.03 0.90 0.94 0.96 0.98 1.01 1931 1
mu2 1.97 0.00 0.02 1.93 1.95 1.97 1.98 2.01 1842 1
gamma1 10.86 0.02 0.74 9.43 10.36 10.85 11.36 12.33 2023 1
gamma2 -37.23 0.04 1.56 -40.34 -38.27 -37.22 -36.18 -34.23 1824 1
sigma1 2.45 0.00 0.26 2.00 2.27 2.44 2.61 3.02 3280 1
sigma2 2.11 0.00 0.22 1.73 1.96 2.09 2.25 2.59 3292 1
lp__ -136.57 0.03 1.83 -141.00 -137.55 -136.26 -135.25 -133.92 5188 1
I have tried translating this to PyMC3 but don’t seem to be getting
very good results. This is my first attempt at PyMC3 so I’m probably
doing something very obviously incorrect.
from pymc3 import Model, Normal, HalfNormal
from pymc3 import NUTS, sample
from pymc3 import Uniform
from pymc3.math import switch
import matplotlib.pyplot as plt
from pymc3 import gelman_rubin
from pymc3 import plot_posterior
v = [x+1 for x in range(100)]
w = [
13.87901,10.81413,13.76257,15.32901,15.84896,15.77714,20.17420,
17.80122,23.23869,19.86830,23.74023,26.80196,20.08339,23.41454,
24.72003,27.33550,26.40307,22.42144,23.87502,32.77224,30.35606,
28.25925,32.63897,36.55082,38.97991,35.09601,36.45973,34.29736,
39.96620,38.65601,41.95645,43.48016,45.17372,42.72125,46.06041,
42.39428,45.35264,46.21309,43.93016,50.07586,51.43260,51.24178,
54.59214,52.47392,52.12661,56.90892,55.29607,61.03261,58.09396,
61.37686,62.70824,62.27555,69.46660,69.41438,70.19747,72.60841,
75.49444,76.19763,71.41520,80.62674,81.19208,84.40751,87.28001,
91.07942,88.39996,94.86559,94.73887,98.28471,100.02560,101.58593,
99.70514,103.80159,107.37174,105.90225,108.80578,113.27819,115.68999,
117.02029,116.05129,117.58048,125.32796,124.56743,126.19457,127.73403,
127.37248,133.34639,133.52229,135.59794,140.05336,141.80790,145.06266,
142.95242,147.43077,151.06044,147.55626,150.10626,151.51017,152.78973,
158.17596,161.43705
]
chg_model = Model()
with chg_model:
# Priors for unknown model parameters
alpha1 = Normal('alpha1', mu=0, sd=10)
alpha2 = Normal('alpha2', mu=0, sd=10)
beta1 = Normal( 'beta1', mu=0, sd=10)
beta2 = Normal( 'beta2', mu=0, sd=10)
sigma1 = HalfNormal('sigma1', sd=10)
sigma2 = HalfNormal('sigma2', sd=10)
tau = Uniform('tau', lower=0, upper=len(w) + 1)
alpha = switch(tau >= v, alpha1, alpha2)
beta = switch(tau >= v, beta1, beta2)
sigma = switch(tau >= v, sigma1, sigma2)
# Expected value of outcome
mu = alpha + beta * v
# Likelihood (sampling distribution) of observations
Y_obs = Normal('Y_obs', mu=mu, sd=sigma, observed=w)
with chg_model:
# draw 500 posterior samples
trace = sample()
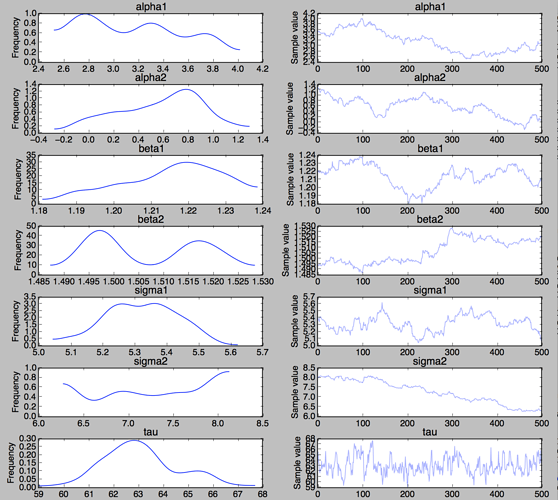
from pymc3 import traceplot
traceplot(trace);
plt.show()
Auto-assigning NUTS sampler...
Initializing NUTS using ADVI...
Average Loss = 367.7: 11% 21718/200000 [00:02<00:23, 7507.08it/s]
Convergence archived at 21800
Interrupted at 21,800 [10%]: Average Loss = 1,453.1
100% 1000/1000 [02:22<00:00, 7.05it/s]/Library/Frameworks/Python.framework/Versions/3.5/lib/python3.5/site-packages/pymc3/step_methods/hmc/nuts.py:448: UserWarning: Chain 0 reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
'reparameterize.' % self._chain_id)