Solution for 1(2?) hierarchical levels
Replaced the data with the samples from the real world:
import xarray as xr
import arviz as az
import pymc3 as pm
import numpy as np
# Create xarray with test data
coords = {'exercise': ['Squat', 'Overhead Press', 'Deadlift'],
'date': ['2020-02-24', '2020-02-26', '2020-03-21', '2020-03-25', '2020-04-06', '2020-07-25', '2020-08-10'],
'observation': [1, 2, 3, 4, 5, 6]}
dims = coords.keys()
velocities = [[[1.198, 1.042, 0.875, 0.735, 0.574, 0.552],
[1.261, 0.993, 0.857, 0.828, 0.624, 0.599],
[1.224, 1.028, 0.990, 0.742, 0.595, 0.427],
[1.112, 0.999, 0.865, 0.751, 0.607, 0.404],
[1.157, 1.010, 0.849, 0.716, 0.593, 0.536],
[1.179, 1.060, 0.898, 0.783, 0.615, 0.501],
[1.209, 1.192, 0.979, 1.025, 0.875, 0.887]],
[[0.911, 0.933, 0.779, 0.629, 0.528, 0.409],
[1.004, 0.863, 0.770, 0.611, 0.511, 0.376],
[0.980, 0.828, 0.766, 0.611, 0.529, 0.349],
[1.024, 0.933, 0.841, 0.734, 0.551, 0.287],
[0.985, 0.852, 0.599, 0.522, 0.313, 0.234],
[0.996, 0.763, 0.758, 0.542, 0.463, 0.378],
[1.066, 0.915, 0.786, 0.686, 0.565, 0.429]],
[[0.654, 1.004, 0.727, 0.483, 0.344, 0.317],
[0.885, 0.738, 0.577, 0.495, 0.335, 0.291],
[0.749, 0.695, 0.539, 0.461, 0.395, 0.279],
[0.801, 0.548, 0.404, 0.424, 0.337, 0.338],
[0.770, 0.642, 0.602, 0.493, 0.394, 0.290],
[0.766, 0.545, 0.426, 0.431, 0.349, 0.329],
[0.787, 0.640, 0.480, 0.401, 0.395, 0.304]]]
loads = [[[20.0, 40.0, 60.0, 80.0, 100.0, 110.0],
[20.0, 40.0, 60.0, 80.0, 100.0, 110.0],
[20.0, 40.0, 60.0, 80.0, 100.0, 117.5],
[20.0, 40.0, 60.0, 80.0, 100.0, 115.0],
[20.0, 40.0, 60.0, 80.0, 100.0, 110.0],
[20.0, 40.0, 60.0, 80.0, 100.0, 110.0],
[20.0, 30.0, 40.0, 50.0, 60.0, 70.0]],
[[20.0, 25.0, 30.0, 35.0, 40.0, 45.0],
[20.0, 25.0, 30.0, 35.0, 40.0, 45.0],
[20.0, 25.0, 30.0, 35.0, 40.0, 45.0],
[20.0, 25.0, 30.0, 35.0, 40.0, 45.0],
[20.0, 30.0, 40.0, 45.0, 50.0, 52.5],
[20.0, 30.0, 35.0, 40.0, 42.5, 45.0],
[20.0, 25.0, 30.0, 35.0, 40.0, 45.0]],
[[60.0, 80.0, 100.0, 120.0, 140.0, 145.0],
[60.0, 80.0, 100.0, 120.0, 140.0, 145.0],
[60.0, 80.0, 100.0, 120.0, 127.5, 140.0],
[60.0, 100.0, 120.0, 125.0, 140.0, 145.0],
[60.0, 80.0, 100.0, 120.0, 130.0, 140.0],
[60.0, 100.0, 120.0, 125.0, 130.0, 132.5],
[70.0, 90.0, 120.0, 135.0, 140.0, 150.0]]]
dataset = xr.Dataset({'velocity': (dims, velocities),
'load': (dims, loads)},
coords = coords)
dataset['velocity_std'] = (dataset['velocity'] - dataset['velocity'].mean())/dataset['velocity'].std()
dataset['load_std'] = (dataset['load'] - dataset['load'].mean())/dataset['load'].std()
dataset
New model:
# Create PyMC
n_exercises = dataset.dims['exercise']
n_dates = dataset.dims['date']
n_observations = dataset.dims['observation']
exercise_idx = [[[i]*n_observations for j in np.arange(n_dates)] for i in np.arange(n_exercises)]
velocity = dataset['velocity_std']
load = dataset['load_std']
with pm.Model(coords = coords) as model:
# Inputs
exercise_idx_shared = pm.Data('exercise_idx', exercise_idx, dims = dims)
velocity_shared = pm.Data('velocity', velocity, dims = dims)
load_shared = pm.Data('load', load, dims = dims)
# Global parameters
global_intercept = pm.Normal('global_intercept', mu = 0, sigma = 1)
global_intercept_sd = pm.Exponential('global_intercept_sd', lam = 1)
global_slope = pm.Normal('global_slope', mu = 0, sigma = 1)
global_slope_sd = pm.Exponential('global_slope_sd', lam = 1)
# Exercise parameters
exercise_intercept = pm.Normal('exercise_intercept', mu = global_intercept, sigma = global_intercept_sd, dims = 'exercise')
exercise_slope = pm.Normal('exercise_slope', mu = global_slope, sigma = global_slope_sd, dims = 'exercise')
# Model error
sigma = pm.Exponential('sigma', lam = 1)
# Expected value
mu = exercise_intercept[exercise_idx_shared] + exercise_slope[exercise_idx_shared]*velocity_shared
# Likelihood of observed values
likelihood = pm.Normal('likelihood',
mu = mu,
sigma = sigma,
observed = load_shared,
dims = dims)
As I understand, the trick was to get the exercise_idx down to the lowest level in the matrix:
exercise_idx = [[[i]*n_observations for j in np.arange(n_dates)] for i in np.arange(n_exercises)]
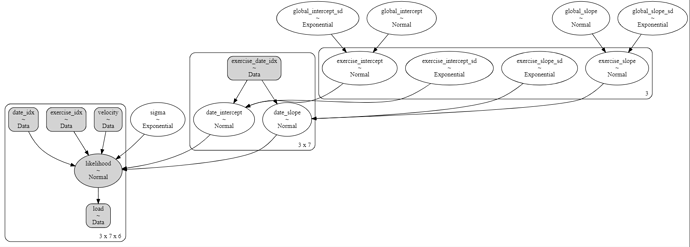
Solution for 2(3?) level hierarchy
The key was once again the indexing. In addition to exercise_idx, the equivivalent for date_idx was needed, and the intermediate indexing exercise_date_idx. The first ones are the full array with the values replaced by the index number of the corresponding coord. The intermediate index list brings the index from the upper (exercise) coord down to the current (date) coord.
# Create PyMC
n_exercises = dataset.dims['exercise']
n_dates = dataset.dims['date']
n_observations = dataset.dims['observation']
exercise_date_idx = [[i]*n_dates for i in np.arange(n_exercises)]
exercise_idx = [[[i]*n_observations for j in np.arange(n_dates)] for i in np.arange(n_exercises)]
date_idx = [[[j]*n_observations for j in np.arange(n_dates)] for i in np.arange(n_exercises)]
velocity = dataset['velocity_std']
load = dataset['load_std']
with pm.Model(coords = coords) as model:
# Inputs
exercise_date_idx_shared = pm.Data('exercise_date_idx', exercise_date_idx, dims = ['exercise', 'date'])
exercise_idx_shared = pm.Data('exercise_idx', exercise_idx, dims = dims)
date_idx_shared = pm.Data('date_idx', date_idx, dims = dims)
velocity_shared = pm.Data('velocity', velocity, dims = dims)
load_shared = pm.Data('load', load, dims = dims)
# Global parameters
global_intercept = pm.Normal('global_intercept', mu = 0, sigma = 1)
global_intercept_sd = pm.Exponential('global_intercept_sd', lam = 1)
global_slope = pm.Normal('global_slope', mu = 0, sigma = 1)
global_slope_sd = pm.Exponential('global_slope_sd', lam = 1)
# Exercise parameters
exercise_intercept = pm.Normal('exercise_intercept', mu = global_intercept, sigma = global_intercept_sd, dims = 'exercise')
exercise_intercept_sd = pm.Exponential('exercise_intercept_sd', lam = 1, dims = 'exercise')
exercise_slope = pm.Normal('exercise_slope', mu = global_slope, sigma = global_slope_sd, dims = 'exercise')
exercise_slope_sd = pm.Exponential('exercise_slope_sd', lam = 1, dims = 'exercise')
# Date parameters
date_intercept = pm.Normal('date_intercept', mu = exercise_intercept[exercise_date_idx_shared], sigma = exercise_intercept_sd[exercise_date_idx_shared], dims = ['exercise', 'date'])
date_slope = pm.Normal('date_slope', mu = exercise_slope[exercise_date_idx_shared], sigma = exercise_slope_sd[exercise_date_idx_shared], dims = ['exercise', 'date'])
# Model error
sigma = pm.Exponential('sigma', lam = 1)
# Expected value
mu = date_intercept[exercise_idx_shared, date_idx_shared] + date_slope[exercise_idx_shared, date_idx_shared]*velocity_shared
# Likelihood of observed values
likelihood = pm.Normal('likelihood',
mu = mu,
sigma = sigma,
observed = load_shared,
dims = dims)