I’m currently trying to implement an errors-in-variables model (a regression model that accounts for errors in the independent variables) in PyMC3.
In a recently published paper (https://arxiv.org/pdf/1906.03989.pdf) I’ve read that
"Except for the simplest case of linear regression [29], EIV modeling almost always
requires auxiliary information or data to correct the mismeasurement bias in estimation.
[…] However, without additional data, Bayesian EIV
modeling is currently the most powerful and flexible approach, as it allows incorporating
additional information in the form of distributional assumptions [15].
So let’s say we have a simple linear errors-in-variables model:
where α and β are the parameters of interest, εt and ηt are the errors and xt* denotes the true but unobserved regressor.
First I create some dummy data and add some error:
import numpy as np
intercept = 0
gradient = 2.5
size = 10000
x_true = np.array(np.linspace(0,1,size))
y_true = intercept + gradient * x_true
# adding noise to data
x = x_true + np.random.normal(loc=0, scale=0.5, size=size)
y = y_true + np.random.normal(loc=0, scale=0.5, size=size)
And then I created my model (reproduced it from Using random variables as `observed` · Issue #2226 · pymc-devs/pymc · GitHub and made some adjustments), where I used high sd’s to get noninformative priors and modeled the “true” regressor x* as a random variable, independend from the measurement error:
import pymc3 as pm
with pm.Model() as eiv_model:
# Define priors
intercept = pm.Normal('intercept', mu=0, sd=20)
gradient = pm.Normal('gradient', mu=0, sd=20)
sigma_x = pm.HalfNormal('sigma_x', sd=20)
sigma_y = pm.HalfNormal('sigma_y', sd=20)
true_x = pm.Normal('true_x', mu=0, sd=10)
likelihood_x = pm.Normal('x', mu=true_x, sd=sigma_x, observed=x)
true_y = pm.Deterministic('true_y', intercept + gradient * true_x)
likelihood_y = pm.Normal('y', mu=true_y, sd=sigma_y, observed=y)
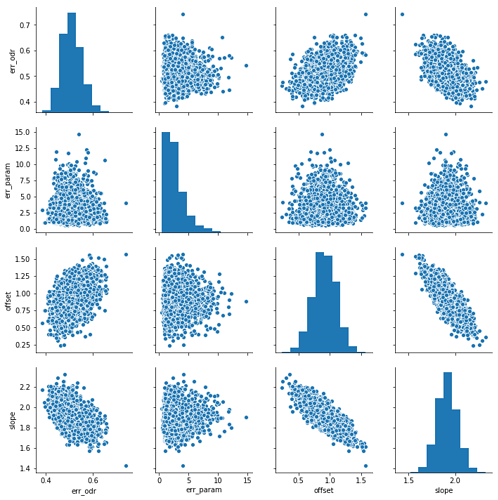
As a result I get:
And now I have some questions:
- Is this model a representative Bayesian errors-in-variables model or is there some bias I’m just not seeing?
- The inferences of the errors, true_x and true_y seems reasonable, but the uncertainty of the intercept and gradient is quite big. Is there a valid way to reduce that uncertainty if all that’s given are the noisy measurements of x and y?
- In the model from Using random variables as `observed` · Issue #2226 · pymc-devs/pymc · GitHub, the shape paramter is used for modelling the true regressor x, but this is only necessary for multivariate models, right?
- Is it possible to modify the model so, that the result corresponds to an orthogonal distance regression model? How would the priors be formulated then?
I’m thankful for any advice, since I’m new to Bayesian inference and probabilistic programming.