I think its easiest to think of errors-in-variables as a repeated-measures model, and then take the number of replicates to be 1 so that:
x_{t,i} \sim N(x_t, \sigma^2_x)
y_{t,i} \sim N(\alpha + \beta x_t, \sigma^2_y)
Then x_t should be a vector and not, as in your case, a scalar, so add shape=x.shape[0] to true_x.
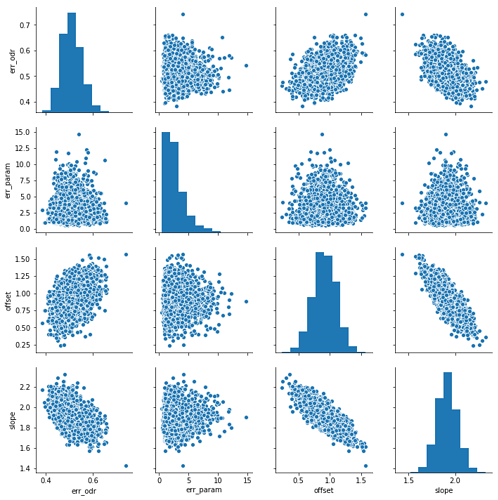
This explains why the intercept and gradient do not differ much from their prior. As true_x was a scalar, it basically estimates the mean and standard deviation of your data (x), so the effective number of samples you’re using is 1 instead of 10000.
With multiple replicates, you have a good amount of information to constrain the variance \sigma_x^2. When you take the number of replicates to 1, you lose the ability to decompose the residual variance into \sigma_x^2 and \sigma_y^2. This is a consequence of the fact that the model is underdetermined; so with this parametrization you should expect divergences to occur as a matter of course.
ODR is a re-parameterization under an assumed variance ratio (\sigma_y^2 = c \cdot \sigma_x^2; c=1 corresponds to ODR, c \neq 1 to weighted ODR). Thus, for classical ODR, the only change is to replace sigma_x and sigma_y with a single sigma_odr.
One final note: Because the variance of your x data is large compared to the range, there is a lot of wiggle room between the slope and the intercept, so even though the true value of the intercept is 0, the marginal mean winds up being close to 1.
Increasing the range of x to np.linspace(0, 10, size) results in a better ability to partition between the slope and intercept terms.
with pm.Model() as model:
err_odr = pm.HalfNormal('err_odr', 5.)
err_param = pm.HalfNormal('err_param', 5.)
x_lat = pm.Normal('x_mu', 0, 5., shape=x.shape[0])
x_obs = pm.Normal('x_obs', mu=x_lat, sd=err_odr, observed=x, shape=x.shape[0])
offset = pm.Normal('offset', 0, err_param)
slope = pm.Normal('slope', 0, err_param)
y_pred = pm.Deterministic('y_pred', offset + slope * x_lat)
y_obs = pm.Normal('y', mu=y_pred, sd=err_odr, observed=y)
trace = pm.sample(800, tune=1500, chains=4, cores=3, init='jitter+adapt_diag')