I am trying to estimate a survival curve with cure fraction.
I would like to use the censor distribution function. However, I don’t know how to work the cure fraction into the analysis.
S = p + (1-p) * Su (where p is the percent who are cured).
![]()
![]()
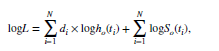
I did an example where I entered the mathematical equations for an exponential distributions. However, I would rather use the censored distributions. I don’t see how you get a statistical function like for instance in excel where you enter the parameters, the x value, and it returns a value either from the pdf or cdf. I’m having trouble figuring how to do this in PyMC.
Below is working code for a model with RFS and PFS for two arms and two treatments.
# Helper functions
def Su(lambda_,t):
return pm.math.exp(-lambda_ * t)
def So(lambda_,t,p):
return p + (1-p)*Su(lambda_,t)
def fu (lambda_,t):
return lambda_*pm.math.exp(-lambda_ * t)
def ho (lambda_,t,p):
return (1-p)*fu(lambda_,t) / (p + (1-p)*Su(lambda_,t))
# Data
dataSOC=osrfs.query('treatment=="SOC"')
T_OS_SOC=dataSOC["os"].to_numpy()
E_OS_SOC=dataSOC["os_event"].to_numpy()
T_RFS_SOC=dataSOC["rfs"].to_numpy()
E_RFS_SOC=dataSOC["rfs_event"].to_numpy()
dataFLIN=osrfs.query('treatment=="SOC FLIN"')
T_OS_FLIN=dataFLIN["os"].to_numpy()
E_OS_FLIN=dataFLIN["os_event"].to_numpy()
T_RFS_FLIN=dataFLIN["rfs"].to_numpy()
E_RFS_FLIN=dataFLIN["rfs_event"].to_numpy()
K=2
J=2
coords={'endpoints': ['OS','RFS'], 'treatments': ['SOC', 'SOC FLIN']}
# Model
from pymc.math import exp, log
with pm.Model() as model_2:
# p global
p_global=pm.Beta("p_global",alpha=1,beta=1)
beta_global=pm.Deterministic("beta_global",pm.math.logit(p_global))
# Priors by treatment
vk=pm.Normal("vk",mu=beta_global, sigma=0.2,shape=(K,1))
sk=pm.HalfNormal("sk",2.5,shape=(K,1))
lind_SOC=pm.Normal("lind_SOC",-1.0,sigma=5);
lind_FLIN=pm.Normal("lind_FLIN",-1.0,sigma=5);
d_SOC=pm.Deterministic("d_SOC",pm.math.invlogit(lind_SOC))
d_FLIN=pm.Deterministic("d_FLIN",pm.math.invlogit(lind_FLIN))
linp_SOC=pm.Normal("linp_SOC",vk[0],sk[0])
p_SOC=pm.Deterministic("p_SOC",pm.math.invlogit(linp_SOC))
linp_FLIN=pm.Normal("linp_FLIN",vk[1],sk[1])
p_FLIN=pm.Deterministic("p_FLIN",pm.math.invlogit(linp_FLIN))
beta0_OS_SOC=pm.Normal("beta0_OS_SOC",mu=0,sigma=100)
lambda_OS_SOC=pm.math.exp(-(beta0_OS_SOC))
OS_SOC = pm.Potential("OS_SOC",E_OS_SOC*log(ho(lambda_OS_SOC,T_OS_SOC,(p_SOC+d_SOC)))+log(So(lambda_OS_SOC,T_OS_SOC,(p_SOC+d_SOC))))
beta0_OS_FLIN=pm.Normal("beta0_OS_FLIN",mu=0,sigma=100)
lambda_OS_FLIN=pm.math.exp(-(beta0_OS_FLIN))
OS_FLIN = pm.Potential("OS_FLIN",E_OS_FLIN*log(ho(lambda_OS_FLIN,T_OS_FLIN,(p_FLIN+d_FLIN)))+log(So(lambda_OS_FLIN,T_OS_FLIN,(p_FLIN+d_FLIN))))
beta0_RFS_SOC=pm.Normal("beta0_RFS_SOC",mu=0,sigma=100)
lambda_RFS_SOC=pm.math.exp(-(beta0_RFS_SOC))
RFS_SOC = pm.Potential("RFS_SOC",E_RFS_SOC*log(ho(lambda_RFS_SOC,T_RFS_SOC,p_SOC))+log(So(lambda_RFS_SOC,T_RFS_SOC,p_SOC)))
beta0_RFS_FLIN=pm.Normal("beta0_RFS_FLIN",mu=0,sigma=100)
lambda_RFS_FLIN=pm.math.exp(-(beta0_RFS_FLIN))
RFS_FLIN = pm.Potential("RFS_FLIN",E_RFS_FLIN*log(ho(lambda_RFS_FLIN,T_RFS_FLIN,p_FLIN))+log(So(lambda_RFS_FLIN,T_RFS_FLIN,p_FLIN)))
data = pm.sample(draws=10000,tune=5000,nuts_sampler="nutpie",cores=20,target_accept=0.99)