As part of a larger model, I’m trying to use the Latent GP implementation in an equivalent way to the Marginal GP implementation (i.e., white noise and Normal observation likelihood), but my attempt yields different results.
Imports
from scipy.spatial.distance import pdist
import pymc3 as pm
import numpy as np
import matplotlib.pyplot as plt
SEED=2020
rng = np.random.default_rng(SEED)
Toy Data
X = np.linspace(0.1,1,20)[:,None]
X = np.vstack([X,X+1.5])
X_ = X.flatten()
y = np.sin(2*np.pi*X_) + rng.normal(0,0.1,len(X_))
σ_fun = (0.1+2*(X_%1.5))/3
y_err = rng.lognormal(np.log(σ_fun),0.1)
y_obs_ = rng.normal(y,y_err,size=(5,len(y)))
y = y_obs_.mean(axis=0)
y_err = y_obs_.std(axis=0)
y_obs = y_obs_.T.flatten()
X_obs = np.tile(X.T,(5,1)).T.reshape(-1,1)
X_obs_ = X_obs.flatten()
idx = np.tile(np.array([i for i,_ in enumerate(X_)]),(5,1)).T.flatten()
Xnew = np.linspace(-0.25,3.25,100)[:,None]
Xnew_ = Xnew.flatten()
ynew = np.sin(2*np.pi*Xnew_)
plt.plot(X,y,'C0o')
plt.errorbar(X_,y,y_err*2,color='C0',ls='none')
plt.plot(X_obs,y_obs,'C1.')
Get lengthscale prior hyperparameters
def get_ℓ_prior(points):
distances = pdist(points[:, None])
distinct = distances != 0
ℓ_l = distances[distinct].min() if sum(distinct) > 0 else 0.1
ℓ_u = distances[distinct].max() if sum(distinct) > 0 else 1
ℓ_σ = max(0.1, (ℓ_u - ℓ_l) / 6)
ℓ_μ = ℓ_l + 3 * ℓ_σ
return ℓ_μ, ℓ_σ
ℓ_μ, ℓ_σ = [stat for stat in get_ℓ_prior(X_)]
Marginal Likelihood Formulation
with pm.Model() as model_marg:
ℓ = pm.InverseGamma('ℓ', mu=ℓ_μ, sigma=ℓ_σ)
η = pm.Gamma('η', alpha=2, beta=1)
cov = η ** 2 * pm.gp.cov.ExpQuad(input_dim=1,ls=ℓ)
gp_marg = pm.gp.Marginal(cov_func=cov)
σ = pm.Exponential('σ', lam=1)
ml_marg = gp_marg.marginal_likelihood('ml_marg', X=X_obs, y=y_obs, noise=σ)
mp_marg = pm.find_MAP()
Sample and Plot Posterior
with model_marg:
mu_marg = gp_marg.conditional("mu_marg", Xnew=Xnew)
samples_marg = pm.sample_posterior_predictive([mp_marg], var_names=['mu_marg'], samples=500)
mu_marg = samples_marg["mu_marg"].mean(axis=0)
sd_marg = samples_marg["mu_marg"].std(axis=0)
plt.plot(Xnew_,mu_marg,'C0')
plt.fill_between(Xnew_,mu_marg+2*sd_marg,mu_marg-2*sd_marg,facecolor='C0',alpha=0.5)
plt.plot(X_obs,y_obs,'C1.')
plt.title('"Marginal" GP Formulation')
MAP values
- ℓ: 0.43
- η: 2.51
- σ: 0.48
Latent Formulation
with pm.Model() as model_lat:
ℓ = pm.InverseGamma('ℓ', mu=ℓ_μ, sigma=ℓ_σ)
η = pm.Gamma('η', alpha=2, beta=1)
σ = pm.Exponential('σ', lam=1)
cov = η ** 2 * pm.gp.cov.ExpQuad(input_dim=1,ls=ℓ) + pm.gp.cov.WhiteNoise(sigma=0.001)
gp_lat = pm.gp.Latent(cov_func=cov)
mu = gp_lat.prior('mu', X=X_obs)
lik_lat = pm.Normal('lik_lat', mu=mu, sd=σ, observed=y_obs)
mp_lat = pm.find_MAP()
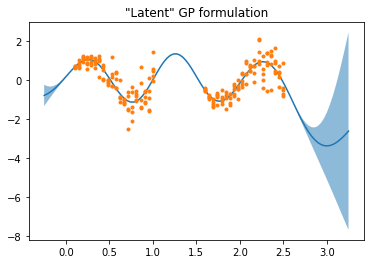
Sample and Plot Posterior
with model_lat:
mu_lat = gp_lat.conditional("mu_lat", Xnew=Xnew)
samples_lat = pm.sample_posterior_predictive([mp_lat], var_names=['mu_lat'], samples=500)
mu_lat = rep_samples["mu_lat"].mean(axis=0)
sd_lat = rep_samples["mu_lat"].std(axis=0)
plt.plot(Xnew_,mu_rep,'C0')
plt.fill_between(Xnew_,mu_lat+2*sd_lat,mu_lat-2*sd_lat,facecolor='C0',alpha=0.5)
plt.plot(X_obs,y_obs,'C1.')
plt.title('"Marginal" GP Formulation')
MAP values:
- ℓ: 0.46
- η: 5.21
- σ: 0.47
Interestingly, the MAP value on the variance scale η is very different between the two. The Marginal GP behaves more like I’d expect, with epistemic uncertainty growing rapidly outside the regions with observations. Any idea how I construct an equivalent to Marginal using the Latent impementation?