I am looking into pymc3 package where I was interested in implementing the package in a scenario where I have several different signals and each signal have different amplitudes.
However, I was stuck in what type of priors I would need to use in order to implement pymc3 into it and likelihood distribution to implement. Scenario example is shown in the following picture

I tried to implement it here, but, every time I keep on getting bad initial energy. Error message can be found here:
pymc3.exceptions.SamplingError: Bad initial energy
Find my portion of the code below:
## Signal 1:
with pm.Model() as model:
# Parameters:
# Prior Distributions:
# BoundedNormal = pm.Bound(pm.Exponential, lower=0.0, upper=np.inf)
# c = BoundedNormal('c', lam=10)
# c = pm.Uniform('c', lower=0, upper=300)
alpha = pm.Normal('alpha', mu = 0, sd = 10)
beta = pm.Normal('beta', mu = 0, sd = 1)
sigma = pm.HalfNormal('sigma', sd = 1)
mu = pm.Normal('mu', mu=0, sigma=1)
sd = pm.HalfNormal('sd', sigma=1)
# Observed data is from a Multinomial distribution:
# Likelihood distributions:
# bradford = pm.DensityDist('observed_data', logp=bradford_logp, observed=dict(value=S1, loc=mu, scale=sd, c=c))
# observed_data = pm.Beta('observed_data', mu=mu, sd=sd, observed=S1)
observed_data = pm.Beta('observed_data', alpha=alpha, beta=beta, mu=mu, sd=sd, observed=S1)
with model:
# obtain starting values via MAP
startvals = pm.find_MAP(model=model)
# instantiate sampler
# step = pm.Metropolis()
step = pm.HamiltonianMC()
# step = pm.NUTS()
# draw 5000 posterior samples
trace = pm.sample(start=startvals, draws=1000, step=step, tune=500, chains=4, cores=1, discard_tuned_samples=True)
# Obtaining Posterior Predictive Sampling:
post_pred = pm.sample_posterior_predictive(trace, samples=500)
print(post_pred['observed_data'].shape)
plt.title('Trace Plot of Signal 1')
pm.traceplot(trace, var_names=['mu', 'sd'], divergences=None, combined=True)
plt.show(block=False)
plt.pause(5) # Pauses the program for 5 seconds
plt.close('all')
pm.plot_posterior(trace, var_names=['mu', 'sd'])
plt.title('Posterior Plot of Signal 1')
plt.show(block=False)
plt.pause(5) # Pauses the program for 5 seconds
plt.close('all')
Edit 1:
I managed to solve the problem with the following code:
with pm.Model() as model:
# Prior Distributions for unknown model parameters:
alpha = pm.HalfNormal('alpha', sigma=10)
beta = pm.HalfNormal('beta', sigma=1)
# Observed data is from a Likelihood distributions (Likelihood (sampling distribution) of observations):
# bradford = pm.DensityDist('observed_data', logp=bradford_logp, observed=dict(value=S1, loc=mu, scale=sd, c=c))
# observed_data = pm.Beta('observed_data', mu=mu, sd=sd, observed=S1)
observed_data = pm.Beta('observed_data', alpha=alpha, beta=beta, observed=S1)
with model:
# obtain starting values via MAP
startvals = pm.find_MAP(model=model)
# instantiate sampler
step = pm.Metropolis()
# step = pm.HamiltonianMC()
# step = pm.NUTS()
# draw 5000 posterior samples
trace = pm.sample(start=startvals, draws=1000, step=step, tune=500, chains=4, cores=1, discard_tuned_samples=True)
# Obtaining Posterior Predictive Sampling:
post_pred = pm.sample_posterior_predictive(trace, samples=500)
print(post_pred['observed_data'].shape)
pm.traceplot(trace, var_names=['alpha', 'beta'], divergences=None, combined=True)
plt.show(block=False)
plt.pause(5) # Pauses the program for 5 seconds
plt.close('all')
pm.plot_posterior(trace, var_names=['alpha', 'beta'])
plt.show(block=False)
plt.pause(5) # Pauses the program for 5 seconds
plt.close('all')
However, I have few probelms:
-
I have multiple files (Say 5 files for each signal) for 5 signals and I get the probability distribution of signals using the Beta distribution from scipy and form a list, then I input the list into the model in Edited code. When I run the code, it runs normally but it reaches a stage where one of the files produces the same error mentioned above. How can I solve this problem?
-
I have tried to reduce the time taken to run each chain by increasing the number of cores, but, when I change the number of cores, it doesn’t run the chains and the program just finishes the code. What could be the reason?
I also have a side question:
Say I would like to implement the bayesian interface in order to see the difference in the pdf of the signals. How do I approach this problem? Do I need to create multiple models and get their posterior distribution? I wanted to like visualise the change of pdf of signal 1 and signal 2 as each single will have different amplitude. Example is shown in the image
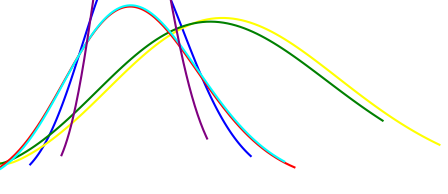
Also, side question, Can I assume that bayesian interface behaves similarly to the concept of the 1D Kalman filter where the bayesian interface can be called bayesian filter?
And what kind of step suitable for this scenario?
 Can you please restate them please? Ideally, can you also post the edited model, the one you say it’s working?
Can you please restate them please? Ideally, can you also post the edited model, the one you say it’s working?