Hello, I’m not sure if this topic had been answered or not, and I’m not familiar with Gaussian Process.
But I am trying to implement Hierarchical Gaussian Process (HGP) based on Hensman, et al (2013). This implementation is implemented in GPy package, and the example notebook can be found here
The idea from the paper is this
which can be extended further as
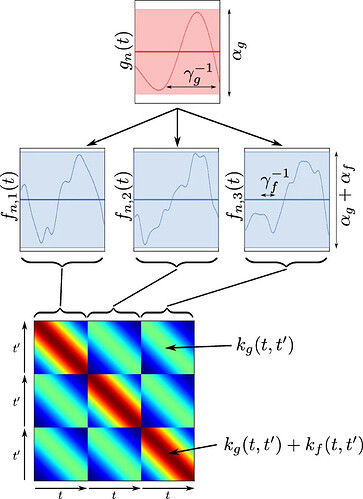
The paper illustrate the model as
where
But as of right now, I don’t really understand PyMC gaussian process enough to implement this. Any guidance is welcome
Thanks!
This is my attempt of following this example, but I think this is still incorrect, and it sample really slow too
Setup + Generate Data
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pymc as pm
%config InlineBackend.figure_format = 'retina'
RANDOM_SEED = 8998
rng = np.random.default_rng(RANDOM_SEED)
az.style.use("arviz-darkgrid")
n_grps = 3
n = 20 # The number of data points
X = np.linspace(0, 10, n_grps * n)[:, None] # The inputs to the GP must be arranged as a column vector
# Define the true covariance function and its parameters
ell_true = 1.0
eta_true = 4.0
cov_func = eta_true**2 * pm.gp.cov.ExpQuad(1, ell_true)
# A mean function that is zero everywhere
mean_func = pm.gp.mean.Zero()
# The latent function values are one sample from a multivariate normal
# Note that we have to call `eval()` because PyMC built on top of Theano
f_true = pm.draw(pm.MvNormal.dist(mu=mean_func(X), cov=cov_func(X)), 1, random_seed=rng)
# The observed data is the latent function plus a small amount of T distributed noise
# The standard deviation of the noise is `sigma`, and the degrees of freedom is `nu`
sigma_true = 1.0
nu_true = 5.0
y = f_true + sigma_true * rng.normal(size=n_grps * n)
indices = np.sort(np.random.permutation(len(y)).reshape(n_grps, n))
X_i = X.reshape(-1)[indices].reshape(n_grps, n, 1)
y_i = y[indices]
## Plot the data and the unobserved latent function
fig = plt.figure(figsize=(10, 4))
ax = fig.gca()
ax.plot(X, f_true, "dodgerblue", lw=3, label="True generating function 'f'")
# ax.plot(X, y, "ok", ms=3, label="Observed data")
for i in range(n_grps):
ax.plot(X_i[i], y_i[i], "o", lw=2, label=f"Partial Obs [{i + 1}]")
ax.set_xlabel("X")
ax.set_ylabel("y")
plt.legend(frameon=True);
Attempt #1
with pm.Model() as model:
ell = pm.Gamma("ell", alpha=2, beta=1)
eta = pm.HalfNormal("eta", sigma=5)
cov_1 = eta**2 * pm.gp.cov.ExpQuad(1, ell)
gp_1 = pm.gp.Latent(cov_func=cov_1)
f_i = [gp_1.prior(f"f_{i}", X=X_i[i]) for i in range(n_grps)]
ell_2 = pm.Gamma("ell_2", alpha=2, beta=1, shape=(n_grps,))
eta_2 = pm.HalfNormal("eta_2", sigma=5, shape=(n_grps,))
conv_2s = [eta_2[i]**2 * pm.gp.cov.ExpQuad(1, ell_2[i]) for i in range(n_grps)]
gp_2s = [pm.gp.Latent(cov_func=conv_2s[i]) for i in range(n_grps)]
g_i = [gp_2s[i].prior(f"g_{i}", X=f_i[i].reshape((X_i.shape[1], 1))) for i in range(n_grps)]
sigma = pm.HalfNormal("sigma", sigma=2.0, shape=(n_grps,))
nu = 1 + pm.Gamma(
"nu", alpha=2, beta=0.1, shape=(n_grps,)
) # add one because student t is undefined for degrees of freedom less than one
y_ = [pm.StudentT(f"y_{i}", mu=g_i[i], lam=(1.0/sigma[i]), nu=nu[i], observed=y_i[i]) for i in range(n_grps)]
idata = pm.sample(200, tune=200, chains=2, cores=6)
Plot
f_posts = az.extract(idata, var_names=[f"f_{i}" for i in range(n_grps)]).transpose("sample", ...)
g_posts = az.extract(idata, var_names=[f"g_{i}" for i in range(n_grps)]).transpose("sample", ...)
# plot the results
fig = plt.figure(figsize=(10, 4))
ax = fig.gca()
color_palette = ["Reds", "Greens", "Blues"]
# for i in range(n_grps):
# plot_gp_dist(ax, f_posts[f"f_{i}"], X_i[i], palette=color_palette[i])
for i in range(n_grps):
plot_gp_dist(ax, g_posts[f"g_{i}"], X_i[i], palette=color_palette[i])
# plot the data and the true latent function
ax.plot(X, f_true, "dodgerblue", lw=3, label="True generating function 'f'")
ax.plot(X, y, "ok", ms=3, label="Observed data")
# axis labels and title
plt.xlabel("X")
plt.ylabel("True f(x)")
plt.title("Posterior distribution over $f(x)$ at the observed values")
plt.legend();
Attempt #2
This version is worse
with pm.Model() as model:
ell = pm.Gamma("ell", alpha=2, beta=1)
eta = pm.HalfNormal("eta", sigma=5)
cov_1 = eta**2 * pm.gp.cov.ExpQuad(1, ell)
gp_1 = pm.gp.Latent(cov_func=cov_1)
f = gp_1.prior("f", X=X_i.reshape(-1, 1))
f_reshape = f.reshape(X_i.shape)
ell_2 = pm.Gamma("ell_2", alpha=2, beta=1, shape=(n_grps,))
eta_2 = pm.HalfNormal("eta_2", sigma=5, shape=(n_grps,))
conv_2s = [eta_2[i]**2 * pm.gp.cov.ExpQuad(1, ell_2[i]) for i in range(n_grps)]
gp_2s = [pm.gp.Latent(cov_func=conv_2s[i]) for i in range(n_grps)]
g_i = [gp_2s[i].prior(f"g_{i}", X=f_reshape[i].reshape((X_i.shape[1], 1))) for i in range(n_grps)]
sigma = pm.HalfNormal("sigma", sigma=2.0, shape=(n_grps,))
nu = 1 + pm.Gamma(
"nu", alpha=2, beta=0.1, shape=(n_grps,)
) # add one because student t is undefined for degrees of freedom less than one
y_ = [pm.StudentT(f"y_{i}", mu=g_i[i], lam=(1.0/sigma[i]), nu=nu[i], observed=y_i[i]) for i in range(n_grps)]
idata = pm.sample(400, tune=400, chains=2, cores=6)