Hi Mister-Knister,
 It looks like you’re fitting a linear mixed model.
It looks like you’re fitting a linear mixed model.
A nice rule of thumb that works for me for pymc3 is to start by writing a model to simulate data – I often find that an observed= can be placed in an obvious place. In this case we’d have
import pymc3 as pm
import theano
import theano.tensor as tt
import numpy as np
import seaborn as sbn
from matplotlib import pyplot as plt
n_treatments = 8
n_groups = 12
print('Total DOF: {} Used DOF: {} Remaining DOF: {}'.format(
n_treatments*n_groups, 2*n_treatments + 2*n_groups + 1,
n_treatments*n_groups - (2*n_treatments + 2*n_groups + 1)
))
var_treat_ = 1 + np.arange(n_treatments, dtype=np.float32)/n_treatments
var_group_ = 1 + np.arange(n_groups, dtype=np.float32)
mean_treat_ = -np.arange(n_treatments, dtype=np.float32)
mean_groups_ = -np.arange(n_groups, dtype=np.float32)
with pm.Model() as model_univar:
mu_t_noise = pm.Normal('_mu_t_noise', 0, 1e-3, shape=n_treatments)
mean_treat = (mu_t_noise + mean_treat_).reshape((n_treatments, 1))
mu_g_noise = pm.Normal('_mu_g_noise', 0, 1e-3, shape=n_groups)
mean_group = (mu_g_noise + mean_groups_).reshape((1, n_groups))
log_sd_t_noise = pm.Normal('_t_noise', 0, 1e-3, shape=n_treatments)
log_sd_g_noise = pm.Normal('_g_noise', 0, 1e-3, shape=n_groups)
sd_treat = tt.exp(log_sd_t_noise + 0.5 * np.log(var_treat_)).reshape((n_treatments, 1))
sd_group = tt.exp(log_sd_g_noise + 0.5 * np.log(var_group_)).reshape((1, n_groups))
sd_noise = 1e-4
mean_mat = mean_treat + mean_group
sd_mat = sd_treat + sd_group + sd_noise
Y_obs = pm.Normal('Y_obs', mu=mean_mat, sd=sd_mat, shape=(n_treatments,n_groups))
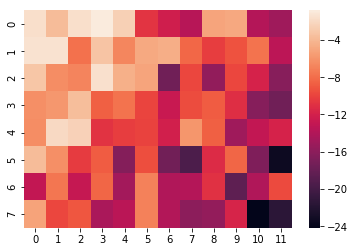
trace = pm.sample_prior_predictive(10)
sbn.heatmap(trace['Y_obs'][0,:,:]);
Total DOF: 96 Used DOF: 41 Remaining DOF: 55
def noncentered_param(basename, shape=None, reshape=None):
names = {'sd': '{}_sd'.format(basename),
'offset': '{}_offset'.format(basename),
'mu': '{}_mu'.format(basename)}
sd = pm.HalfNormal(names['sd'], 1, shape=shape)
mu = pm.Normal(names['mu'], 0, 1, shape=shape)
off = pm.Normal(names['offset'], 0, 1, shape=shape)
var = pm.Deterministic(basename, mu + sd * off)
if reshape:
return var.reshape(reshape)
return var
with pm.Model() as model_univar_inf:
mean_treat = noncentered_param('mean_treat', n_treatments, (n_treatments,1))
mean_group = noncentered_param('mean_group', n_groups, (1, n_groups))
sd_treat = tt.exp(noncentered_param('sd_treat_log', n_treatments, (n_treatments,1)))
sd_group = tt.exp(noncentered_param('sd_group_log', n_groups, (1, n_groups)))
sd_noise = pm.HalfNormal('sd_err', 1.)
mean_mat = mean_treat + mean_group
sd_mat = sd_treat + sd_group + sd_noise
Y_obs = pm.Normal('Y_obs', mu=mean_mat, sd=sd_mat, shape=(n_treatments,n_groups), observed=trace['Y_obs'][0,:,:])
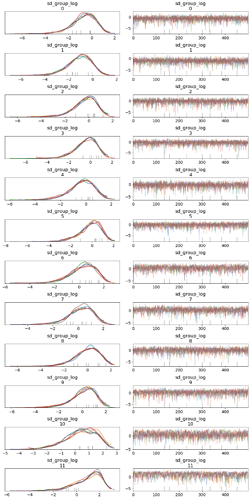
inf_trace = pm.sample(500, tune=1000, chains=6)
pm.traceplot(inf_trace, ['sd_group_log']);
You can also have multiple observations along the 0th axis (though this is not likely to be your use case)
Y_obs = pm.Normal('Y_obs', mu=mean_mat, sd=sd_mat, shape=(n_treatments,n_groups), observed=trace['Y_obs'][:,:,:])
inf_trace = pm.sample(500, tune=1000, chains=6)
When moving to correlations, the typical way is to note that the model above (where sd_mat is explicit) is equivalent (for a single matrix observation) to this model:
with pm.Model() as model_latent_var:
mean_treat = noncentered_param('mean_treat', n_treatments, (n_treatments,1))
mean_group = noncentered_param('mean_group', n_groups, (1, n_groups))
# note that reshaping no longer happens here
sd_treat = tt.exp(noncentered_param('sd_treat_log', n_treatments))
sd_group = tt.exp(noncentered_param('sd_group_log', n_groups))
sd_noise = pm.HalfNormal('sd_err', 1.)
z_treat = pm.Deterministic('z_treat', pm.Normal('_z_treat_offset', 0, 1, shape=n_treatments) * sd_treat).reshape((n_treatments,1))
z_group = pm.Deterministic('z_group', pm.Normal('_z_group_offset', 0, 1, shape=n_groups) * sd_group).reshape((1, n_groups))
mean_mat = (mean_treat + mean_group) + (z_treat + z_group)
Y_obs = pm.Normal('Y_obs', mu=mean_mat, sd=sd_noise, shape=(n_treatments,n_groups), observed=trace['Y_obs'][0,:,:])
inf_trace = pm.sample(500, tune=1000, chains=6)
it is slower (and harder) to sample, as the sampler needs to marginalize over the new z variables; but it gives you more direct control over defining (marginal) correlations between treatments and groups (rows and columns); by allowing you to make more complicated models for z.
If you can define the marginal relationships (row covariance, column covariance) explicitly, take a look at pm.MatrixNormal.
Note, especially, that the “full” covariance for a (N,K) matrix is in fact an (N, K, N, K) tensor because of terms like \mathrm{cor}(X_{1,3},X_{2,8}). In the rare case where you want to test particular structures for that tensor, you still need to unwrap the (N,K) matrix into an (NK, 1) vector, and specify the tensor as the appropriate (NK,NK) matrix.
 It looks like you’re fitting a linear mixed model.
It looks like you’re fitting a linear mixed model.
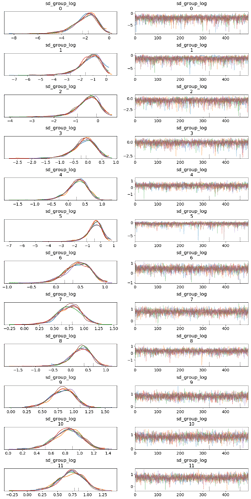
 ] There’s no need to define a custom distribution for this. In fact, you would just get this from
] There’s no need to define a custom distribution for this. In fact, you would just get this from