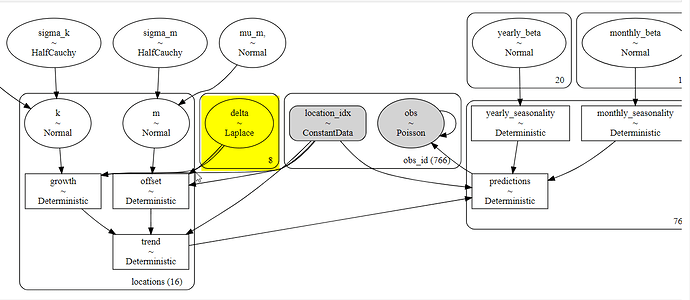
Hello,
I have monthly, sales data for 16 different locations. I built a ‘prophet-like’ model based on @MBrouns video here ( Hierarchical Time Series With Prophet and PyMC3 (Matthijs Brouns) - YouTube
That stated, the model runs fine until I try to make it a hierarchical model with locations. The model samples then errors when it’s transforming the variables with a shape error. Below is the model code and error.
location_idx, locations = pd.factorize(df_sat['location'])
coords_ = {'locations':locations,
'obs_id':np.arange(len(location_idx))}
with pm.Model(coords = coords_) as model:
location_idx_ = pm.ConstantData('location_idx', location_idx, dims = 'obs_id')
mu_k = pm.Normal('mu_k', 0, 100)
sigma_k = pm.HalfCauchy('sigma_k', 5)
k = pm.Normal('k', mu = mu_k, sigma = sigma_k, dims = 'locations')
mu_m = pm.Normal('mu_m,', 0, 100)
sigma_m = pm.HalfCauchy('sigma_m', 5)
m = pm.Normal('m', mu=mu_m, sigma = sigma_m, dims = 'locations')
delta = pm.Laplace('delta', 0, 0.1, shape=n_changepoints)
growth = pm.Deterministic('growth', (k[location_idx_] + at.dot(a, delta)), dims = 'locations')
offset = pm.Deterministic('offset', (m[location_idx_] + at.dot(a, -s * delta)), dims = 'locations')
trend = pm.Deterministic('trend', growth[location_idx_] * t + offset[location_idx_], dims = 'locations')
# yearly_mu = pm.Normal('yearly_mu', 0, 1)
# yearly_sigma = pm.HalfCauchy('yearly_sigma', 0, 1)
yearly_beta = pm.Normal('yearly_beta', mu = 0, sigma = 1, shape = n_components*2)
yearly_seasonality = pm.Deterministic('yearly_seasonality',at.dot(yearly_X(t, 365.25/len(t)), yearly_beta))
# monthly_mu = pm.Normal('monthly_mu', 0, 1)
# monthly_sigma = pm.HalfCauchy('monthly_sigma', 0, 1)
monthly_beta = pm.Normal('monthly_beta', mu = 0, sigma=5, shape = monthly_n_components*2)
monthly_seasonality = pm.Deterministic('monthly_seasonality',at.dot(monthly_X(t, 30.5/len(t)), monthly_beta))
predictions = pm.Deterministic('predictions', np.exp(trend[location_idx_] + yearly_seasonality + monthly_seasonality))
pm.Poisson('obs',
mu = predictions,
observed=df_sat['eaches'],
dims = 'obs_id'
)
Error
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/tmp/ipykernel_5309/3004907719.py in <module>
34 )
35
---> 36 trace_locations = pymc.sampling_jax.sample_numpyro_nuts(tune=2000, draws = 2000)
/opt/conda/lib/python3.7/site-packages/pymc/sampling_jax.py in sample_numpyro_nuts(draws, tune, chains, target_accept, random_seed, initvals, model, var_names, progress_bar, keep_untransformed, chain_method, postprocessing_backend, idata_kwargs, nuts_kwargs)
568 dims=dims,
569 attrs=make_attrs(attrs, library=numpyro),
--> 570 **idata_kwargs,
571 )
572
/opt/conda/lib/python3.7/site-packages/arviz/data/io_dict.py in from_dict(posterior, posterior_predictive, predictions, sample_stats, log_likelihood, prior, prior_predictive, sample_stats_prior, observed_data, constant_data, predictions_constant_data, warmup_posterior, warmup_posterior_predictive, warmup_predictions, warmup_log_likelihood, warmup_sample_stats, save_warmup, index_origin, coords, dims, pred_dims, pred_coords, attrs, **kwargs)
457 pred_coords=pred_coords,
458 attrs=attrs,
--> 459 **kwargs,
460 ).to_inference_data()
/opt/conda/lib/python3.7/site-packages/arviz/data/io_dict.py in to_inference_data(self)
333 return InferenceData(
334 **{
--> 335 "posterior": self.posterior_to_xarray(),
336 "sample_stats": self.sample_stats_to_xarray(),
337 "log_likelihood": self.log_likelihood_to_xarray(),
/opt/conda/lib/python3.7/site-packages/arviz/data/base.py in wrapped(cls)
63 if all((getattr(cls, prop_i) is None for prop_i in prop)):
64 return None
---> 65 return func(cls)
66
67 return wrapped
/opt/conda/lib/python3.7/site-packages/arviz/data/io_dict.py in posterior_to_xarray(self)
102 dims=self.dims,
103 attrs=posterior_attrs,
--> 104 index_origin=self.index_origin,
105 ),
106 dict_to_dataset(
/opt/conda/lib/python3.7/site-packages/arviz/data/base.py in dict_to_dataset(data, attrs, library, coords, dims, default_dims, index_origin, skip_event_dims)
312 default_dims=default_dims,
313 index_origin=index_origin,
--> 314 skip_event_dims=skip_event_dims,
315 )
316 return xr.Dataset(data_vars=data_vars, attrs=make_attrs(attrs=attrs, library=library))
/opt/conda/lib/python3.7/site-packages/arviz/data/base.py in numpy_to_data_array(ary, var_name, coords, dims, default_dims, index_origin, skip_event_dims)
252 # filter coords based on the dims
253 coords = {key: xr.IndexVariable((key,), data=np.asarray(coords[key])) for key in dims}
--> 254 return xr.DataArray(ary, coords=coords, dims=dims)
255
256
/opt/conda/lib/python3.7/site-packages/xarray/core/dataarray.py in __init__(self, data, coords, dims, name, attrs, indexes, fastpath)
404 data = _check_data_shape(data, coords, dims)
405 data = as_compatible_data(data)
--> 406 coords, dims = _infer_coords_and_dims(data.shape, coords, dims)
407 variable = Variable(dims, data, attrs, fastpath=True)
408 indexes = dict(
/opt/conda/lib/python3.7/site-packages/xarray/core/dataarray.py in _infer_coords_and_dims(shape, coords, dims)
153 if s != sizes[d]:
154 raise ValueError(
--> 155 f"conflicting sizes for dimension {d!r}: "
156 f"length {sizes[d]} on the data but length {s} on "
157 f"coordinate {k!r}"
ValueError: conflicting sizes for dimension 'locations': length 766 on the data but length 16 on coordinate 'locations'
I’ve tried taking the dims=locations out in various places. Maybe this has to do with the shape of the delta variable but when I try to make shape = (16, n_changepoints), another error pops up. I’m not sure where the shape error is actually coming from.