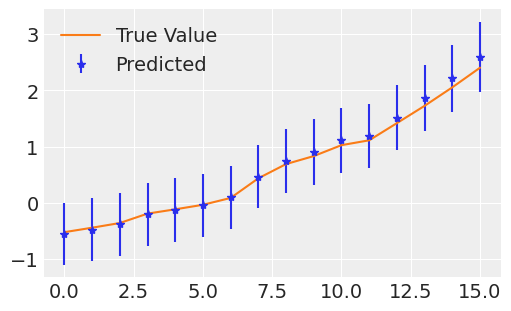
I tried running MCMC, it took around 5 hours to run, but the results are almost the same. Below is the model I am running-
with pm.Model(coords=coords, check_bounds=False) as autoregressive_model:
sigma = pm.HalfNormal("sigma", .2)
ar_init_obs = pm.MutableData("ar_init_obs", np.squeeze(u_scale[0:lags,0:features]), dims=("features"))
ar_init = pm.Normal("ar_init", observed=ar_init_obs, dims=("features"))
PAT_true = pm.MutableData("PAT_obs", PAT_obs, dims=("trials"))
mu2_a = pt.zeros(features)
sd_dist_a = pm.Exponential.dist(1.0, shape=features)
chol2_a, corr_a, stds_a = pm.LKJCholeskyCov('chol_cov_ar', n=features, eta=2,
sd_dist=sd_dist_a, compute_corr=True)
A_ar = pm.MvNormal('A_ar', mu=mu2_a, chol=chol2_a, shape=(features))
sigmas_Q_ar = pm.HalfNormal('sigmas_Q_ar', sigma=1, shape= (features))
Q_ar = pt.diag(sigmas_Q_ar)
intercept = pm.Normal("intercept", mu=0, sigma=1, dims=("features"))
def ar_step(x, A, Q, intercept):
mu = A*x
#mu = pm.math.dot(A,x) + intercept
next_x = mu + pm.MvNormal.dist(mu=0, tau=Q)
return next_x, collect_default_updates(inputs = [x, A, Q, intercept], outputs = [next_x])
ar_innov, ar_innov_updates = pytensor.scan(
fn=ar_step,
outputs_info=[ar_init],
non_sequences=[A_ar, Q_ar, intercept],
n_steps=trials-lags,
strict=True,
)
ar_innov_obs = pm.MutableData("ar_innov_obs", u_scale[lags:,0:features], dims=("steps","features"))
ar_innov = autoregressive_model.register_rv(ar_innov, name="ar_innov", observed=ar_innov_obs, dims=("steps","features"))
ar = pm.Deterministic("ar", pt.concatenate([ar_init.reshape((1,features)), ar_innov], axis=0))
ar_obs = pm.Deterministic("ar_obs", pt.concatenate([ar_init_obs.reshape((1,features)), ar_innov_obs], axis=0))
There are in total 8 features and the graph which I displayed is for one of the features. Is there any issue with the above modeling process?
Below is the distribution of sigmas

Below is the prediction for feature 0