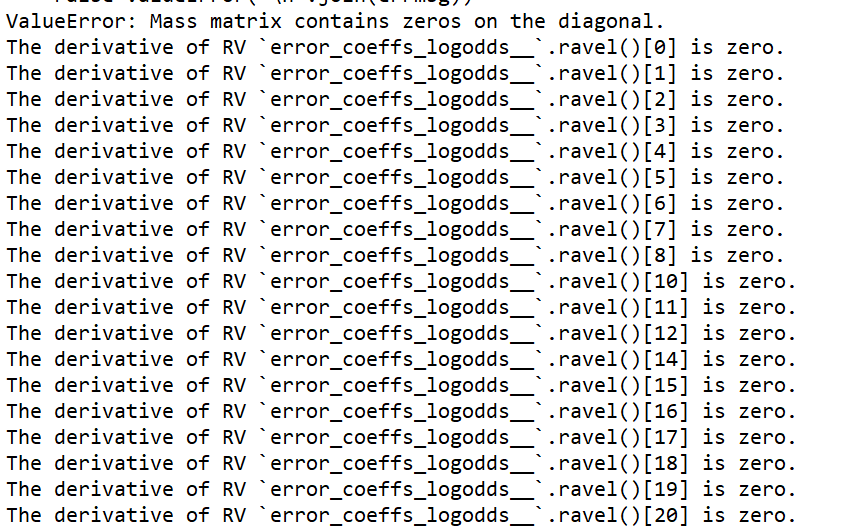
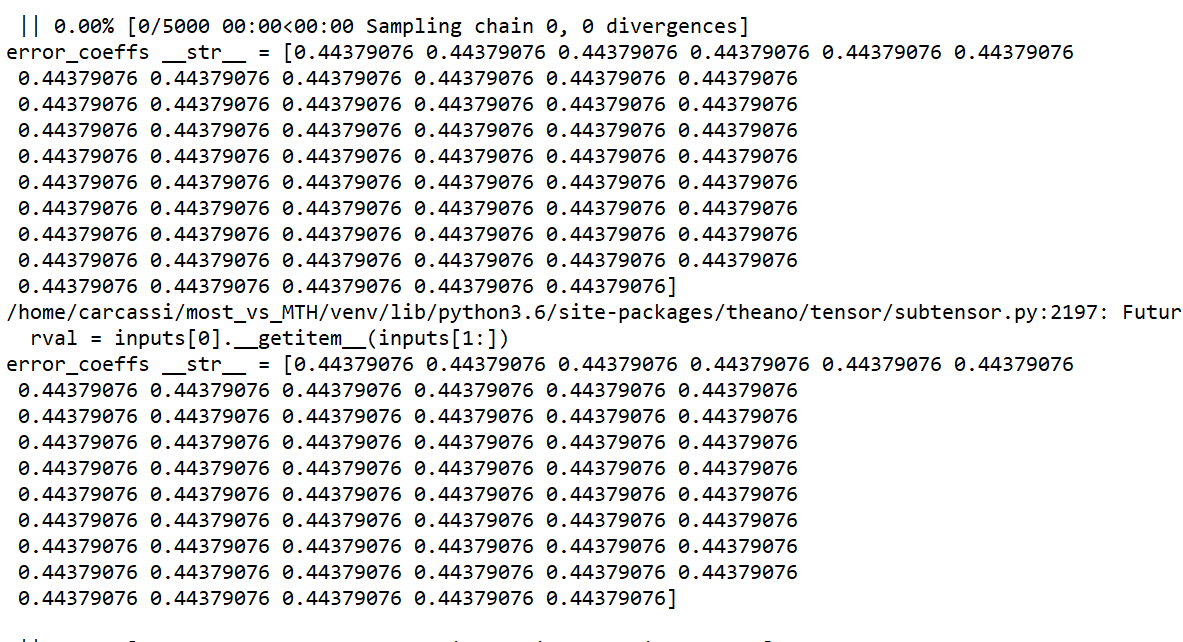
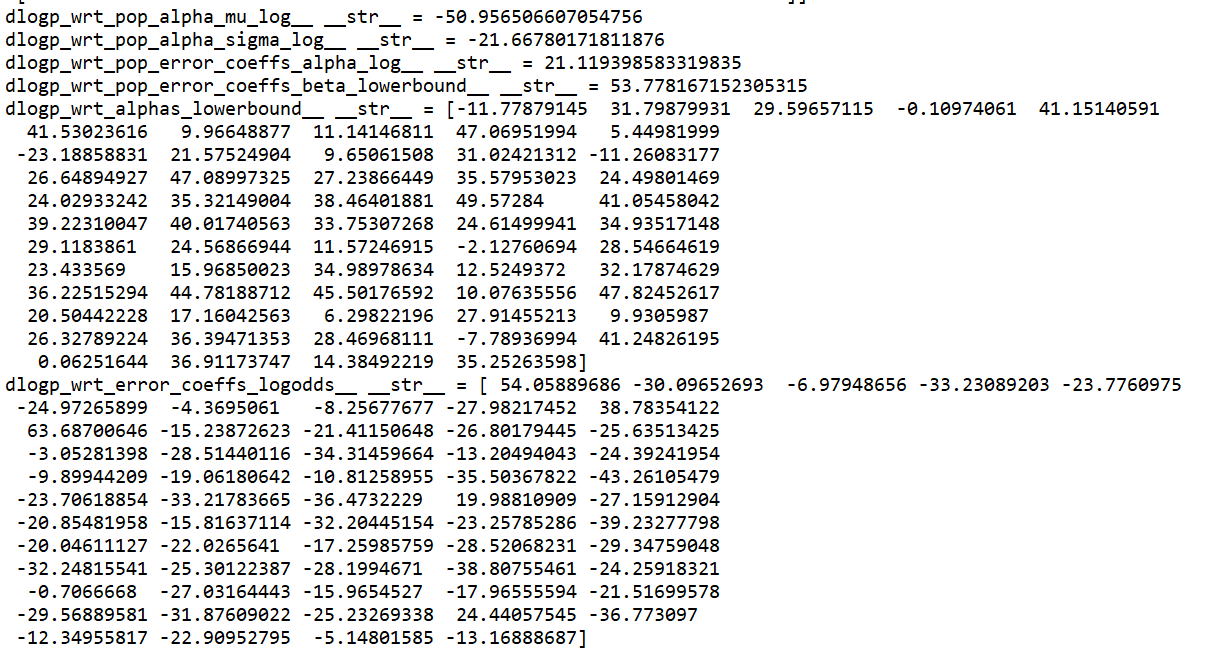
Here is the model with a sample dataset (where the model doesn’t fail however, as it’s much smaller than the actual dataset). It’s quite complicated!
import numpy as np
import pymc3 as pm
import theano as tt
import theano.tensor as T
def create_model(num_participants, num_states, possible_signals_array,
real_signals_indices, types, picked_signals_indices,
picsizes_values, participants_indices, states_values):
with pm.Model() as model:
types = tt.shared(types, name='types')
pop_alpha_mu = pm.HalfNormal(
'pop_alpha_mu', sigma=3)
pop_alpha_sigma = pm.HalfNormal(
'pop_alpha_sigma', sigma=3)
alphas = pm.TruncatedNormal(
"alphas",
mu=pop_alpha_mu,
sigma=pop_alpha_sigma,
lower=0,
shape=num_participants
)
error_coeffs_alpha = pm.HalfNormal(
'pop_error_coeffs_alpha', sigma=1)
error_coeffs_beta = pm.TruncatedNormal(
'pop_error_coeffs_beta', mu=3, sigma=1.5, lower=0)
error_coeffs = pm.Beta(
'error_coeffs',
alpha=error_coeffs_alpha,
beta=error_coeffs_beta,
shape=num_participants
)
considered_signals = (types.dimshuffle(0, 'x') >=
types.dimshuffle('x', 0))
min_picsize = min(picsizes_values)
probability_accept = T.zeros(shape=(len(picsizes_values),
len(real_signals_indices)))
real_signals_indices = tt.shared(real_signals_indices,
name='real_signals_indices')
for index_state, state in enumerate(range(min_picsize, num_states)):
local_poss_sig_array = tt.shared(
possible_signals_array[state], name='local_poss_sig_array'
)
real_signals_array = local_poss_sig_array[real_signals_indices]
unique_alt_profiles, index_signals_profile = T.extra_ops.Unique(
axis=0, return_inverse=True)(considered_signals)
unique_alt_profiles_expanded = T.tile(
unique_alt_profiles[np.newaxis, :,:, np.newaxis],
reps=(alphas.shape[0],1,1,local_poss_sig_array.shape[-1])
)
language_l = local_poss_sig_array / local_poss_sig_array.sum(
axis=-1, keepdims=True)
l0 = language_l[np.newaxis,:,:]
l0_extended = T.tile(l0[:,np.newaxis,:,:],
reps=(1, unique_alt_profiles.shape[0],1,1))
unnorm_s1 = l0_extended ** alphas[
:,np.newaxis, np.newaxis, np.newaxis]
s1_unnorm = T.switch(unique_alt_profiles_expanded, unnorm_s1, 0)
s1 = s1_unnorm / s1_unnorm.sum(axis=-2, keepdims=True)
s1_filled = T.switch(unique_alt_profiles_expanded, s1, 1)
l2 = s1_filled / s1_filled.sum(axis=-1, keepdims=True)
language_possible = l2[:, index_signals_profile,
T.arange(l2.shape[2]), :]
l2_language = language_possible[:,real_signals_indices,:]
unnorm_s3 = l2_language**alphas[:,np.newaxis,np.newaxis]
s3 = unnorm_s3 / unnorm_s3.sum(axis=-2, keepdims=True)
uniform_unnorm = T.ones_like(s3)
uniform = uniform_unnorm / uniform_unnorm.sum(axis=-2, keepdims=True)
error_params_expanded = error_coeffs[:,np.newaxis,np.newaxis]
s3 = error_params_expanded*uniform+(1-error_params_expanded)*s3
relevant_indices = (picsizes_values == state).nonzero()[0]
subtensor = s3[
participants_indices[relevant_indices],:,
states_values[relevant_indices]]
probability_accept = T.set_subtensor(
probability_accept[relevant_indices],
subtensor)
# save the probability of acceptance
pm.Deterministic("probability_accept", probability_accept)
### observed
obs = pm.Categorical(
"picked_signals",
p=probability_accept,
shape=len(picsizes_values),
observed=picked_signals_indices)
step = pm.NUTS(target_accept=0.99)
trace=pm.sample(step=step, cores=1)
return trace
args = {
'num_participants': 3,
'num_states': 4,
'possible_signals_array': [
np.array([[0],[0],[0], [1], [0], [1], [1], [1], [0], [0], [0], [1], [1], [0], [0], [1], [1]]),
np.array([[0, 1], [0, 1], [0, 1], [1, 1], [0, 1], [1, 0], [1, 0], [1, 0], [0, 1], [0, 1], [0, 1], [1, 0], [1, 0], [0, 1], [0, 1], [1, 0], [1, 0]]),
np.array([[0, 0, 1], [0, 0, 1], [0, 0, 1], [0, 1, 0], [0, 1, 1], [1, 0, 0], [1, 0, 0], [1, 0, 0], [0, 1, 1], [0, 1, 1], [0, 0, 1], [1, 0, 0], [1, 1, 0], [0, 1, 1], [0, 0, 1], [1, 0, 0], [1, 1, 0]]),
np.array([[0, 0, 1, 1], [0, 0, 1, 1], [0, 0, 0, 1], [0, 0, 1, 0], [0, 0, 1, 1], [1, 0, 0, 0], [1, 1, 0, 0], [1, 0, 0, 0], [0, 1, 1, 1], [0, 1, 1, 1], [0, 0, 1, 1], [1, 0, 0, 0], [1, 1, 0, 0], [0, 1, 1, 1], [0, 0, 0, 1], [1, 0, 0, 0], [1, 1, 1, 0]])
],
'real_signals_indices': np.array([0, 1, 2, 3, 4, 5, 6, 7, 8]),
'types': np.array([1, 2, 1, 1, 1, 1, 2, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2]),
'picked_signals_indices': np.array([4, 5, 6, 3, 2, 5, 3, 0, 0]),
'picsizes_values': np.array([1, 2, 2, 2, 3, 2, 2, 3, 3]),
'participants_indices': np.array([0, 0, 0, 1, 1, 1, 2, 2, 2]),
'states_values': np.array([1, 2, 1, 1, 3, 0, 2, 3, 0])
}
trace = create_model(**args)
I am wondering if there is any way to check if alpha is overparameterized? Usually I would check if it shows random walk like behaviour in the trace, but in this case there is no trace to look at! Another question: is there any way to print the various logp, dlogp etc for the frogleaps? Thank you so much for the help, I really appreciate it!