I have a dataset that describes the wealth index of Rwandan housholds: wealth.csv (29.0 KB)
Its shape and distribution is:

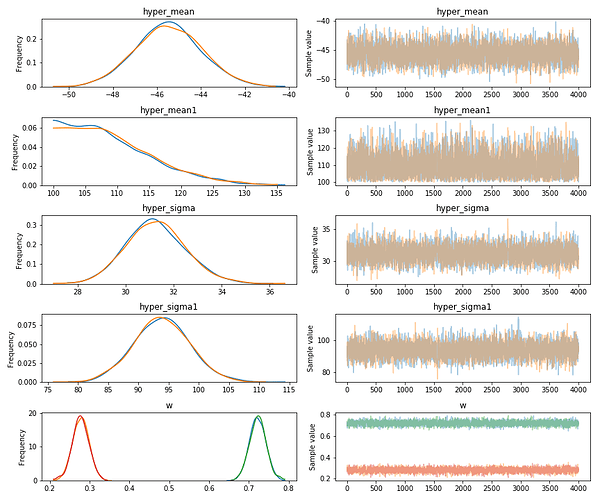
I am using Pymc3 Mixture Modeling to divide wealth index into 2 separate groups (presumably rich and poor).
with pm.Model() as model:
hyper_mean = pm.Uniform('hyper_mean', -100, 10)
hyper_mean1 = pm.Uniform('hyper_mean1', 100, 300)
hyper_sigma = pm.Uniform('hyper_sigma', 0, 100)
hyper_sigma1 = pm.Uniform('hyper_sigma1', 0, 150)
component = pm.Normal.dist(mu=hyper_mean, sd=hyper_sigma)
component1 = pm.Normal.dist(mu=hyper_mean1, sd=hyper_sigma1)
w = pm.Dirichlet('w', a=np.array([1, 1]))
like = pm.Mixture('like', w=w, comp_dists=[component, component1], observed=data)
with model:
trace = pm.sample(5000, tune=2500, njobs=1)[1000:]
As I am uncertain about the prior parameters of Rwandan wealth, I (think I) am using non restrictive priors; thus allowing the data to have a strong influence on the posterior.
Resulting in:
When I visualize posterior values over observed data, the observed data is flatter around the 100-200 wealth index:
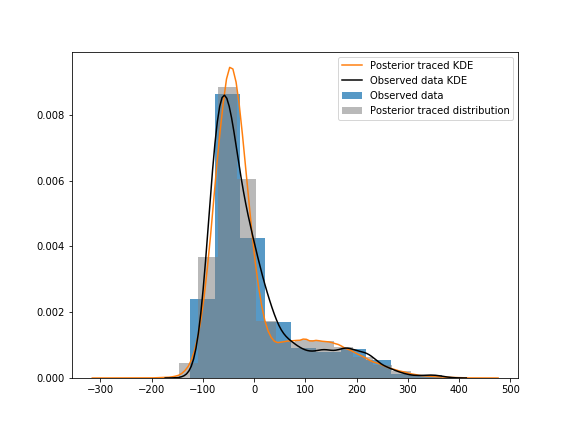
I’ve tried using the Student_T distribution as the components instead of the Normal one, but it gave the model longer tails and a bigger hump around the 100-200 mark.
With the priors being so non restrictive, I would think the data would take over the posterior.
Do you suggest using a different distribution to represent the components, or reparametrize, or do anything else to have the posterior align better?