Hello,
I am trying to implement a dynamical hierarchical structure of layers in a model.
For example:
Imagine you have a Hierarchy of three components: ‘A’, ‘B’, ‘C’. The hierarchy is given by a String with a structure like this:
"""
0->1,2->0
1->2->1
2->3->2,3
"""
In the first line would mean that the component A can only match with component B; 1 or 2 and with component C; 0
So the idea should be that each component in the hierarchy will affect the distribution of each child. So the dist[B;1] will be affected by the dist[A;0].
To implement that idea of dependency I have tried to do it dynamically through a for loop and dependency through the distributions.
- components contains the unique values of each component
- indexes contains the indexes of each level of the hierarchy
niter=1
with self.model:
previous_sigma = pm.Exponential('sgm-hyper-prior', hierarchy_spec.hiperparams[0])
niter = 1
for _ in elements:
if niter==1:
previous_dist = pm.Normal('hie-'+":".join(elements[:niter]),
mu = hierarchy_spec.hiperparams[0],
sigma = self.model.named_vars.get(previous_sigma.name),
shape = len(combinations[":".join(elements[:niter])]))
if niter!=len(elements):
previous_sigma = pm.HalfNormal('sgm-hie-'+":".join(elements[:niter]),
sigma = self.model.named_vars.get(previous_sigma.name),
shape = len(combinations[":".join(elements[:niter])]))
hierarchy_spec.hierarchy_layers.append(previous_sigma.name)
niter+=1
elif(niter==len(elements)):
previous_dist = pm.Normal("hie-"+":".join(elements[:niter]),
mu = self.model.named_vars.get(previous_dist.name)[indexes[":".join(elements[:niter])]],
sigma = self.model.named_vars.get(previous_sigma.name)[indexes[":".join(elements[:niter])]],
shape = len(combinations[":".join(elements[:niter])]))
else:
previous_dist = pm.Normal("hie-"+":".join(elements[:niter]),
mu = self.model.named_vars.get(previous_dist.name)[indexes[":".join(elements[:niter])]],
sigma = self.model.named_vars.get(previous_sigma.name)[indexes[":".join(elements[:niter])]],
shape = len(combinations[":".join(elements[:niter])]))
previous_sigma = pm.HalfNormal('sgm-hie-'+":".join(elements[:niter]),
sigma = self.model.named_vars.get(previous_sigma.name)[indexes[":".join(elements[:niter])]],
shape = len(combinations[":".join(elements[:niter])]))
niter+=1
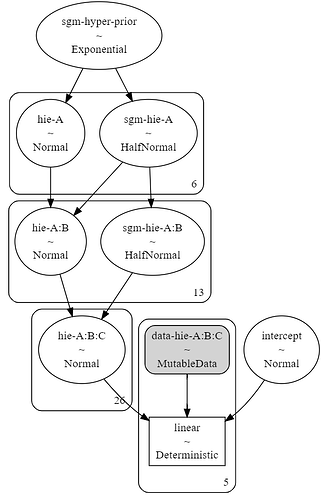
The structure of the model should be something like this and I obtain the correct one:
However when I started sampling the model, PYMC returned the following error:
---------------------------------------------------------------------------
NotImplementedError Traceback (most recent call last)
File c:\Users\O002095\Anaconda3\envs\withprofy\lib\site-packages\pytensor\compile\function\types.py:970, in Function.__call__(self, *args, **kwargs)
968 try:
969 outputs = (
--> 970 self.vm()
971 if output_subset is None
972 else self.vm(output_subset=output_subset)
973 )
974 except Exception:
File c:\Users\O002095\Anaconda3\envs\withprofy\lib\site-packages\pytensor\graph\op.py:543, in Op.make_py_thunk..rval(p, i, o, n, params)
539 @is_thunk_type
540 def rval(
541 p=p, i=node_input_storage, o=node_output_storage, n=node, params=None
542 ):
--> 543 r = p(n, [x[0] for x in i], o)
544 for o in node.outputs:
File c:\Users\O002095\Anaconda3\envs\withprofy\lib\site-packages\pymc\logprob\transforms.py:100, in TransformedVariable.perform(self, node, inputs, outputs)
99 def perform(self, node, inputs, outputs):
--> 100 raise NotImplementedError("These `Op`s should be removed from graphs used for computation.")
If I implement the hierarchy statically and without dynamic indexing through the distributions, everything runs and samples correctly:
- A,B,C contains the unique values of each component
- A_B, B_C… contains the index of the corresponding hierarchical level
with model:
intercept = pm.Normal('intercept', mu=1, sigma = 10)
# pm.MutableData("data-cta_text", X['cta_text'].values)
pm.MutableData("data-C_obs_idx", ctas_obs_idx)
sigma_hyp = pm.Exponential("sigma_hyp", 0.005)
mu_A = pm.Normal("A", mu=0, sigma=sigma_hyp, shape=len(A))
sigma_A = pm.HalfNormal("sigma_A", sigma=sigma_hyp, shape=len(A))
mu_B = pm.Normal("mu_B", mu = mu_A[A_B_idx], sigma=sigma_A[A_B_idx], shape=len(A_B))
sigma_B = pm.HalfNormal('sigma_B', sigma = sigma_micro[A_B_idx], shape=len(A_B))
mu_C = pm.Normal("mu_C", mu=mu_B[B_C_idx], sigma=sigma_B[B_C_idx], shape = len(B_C))
linear = pm.Deterministic('linear', (intercept + mu_C[model.named_vars['data-C_obs_idx']]))
In both cases there is a likelihood used with the same structure:
with mlb.model:
pm.MutableData('target', X_c['target'].values)
pm.Normal("y_hat", mu=mlb.model.named_vars.get('linear'), sigma=1, observed=mlb.model.named_vars.get('target'))
Thanks in advance