Hello!
I would like to examine the traces from fitted time series to see how well they described. When I tried something simple-minded like trace[‘x’], where x is the variable assigned to the time series, I get an “Unknown variable” error. Could someone tell me what I am missing here?
Thanks in advance,
Gregory
It would be helpful if you could show a small example that yields the error that you’re coming across. Otherwise, I’d recommend to make sure that you aren’t accidentally instantiating the model a second time.
Thanks for the suggestion. Here’s is an example:
import numpy as np
import matplotlib.pyplot as plt
import pymc3 as pm
print('Running on PyMC3 v{}'.format(pm.__version__))
T = 1001; # this many samples
y = np.zeros((T,))
for i in range(1,T):
y[i] = 0.95 * y[i-1] + np.random.normal()
delta = np.diff(y) # Get difference series (demeaned)
print(delta)
print(len(delta))
sd = 1.0 # SD of priors for beta
lags = 2 # this many number of lags
with pm.Model() as model:
beta = pm.Normal('beta', mu=0, sd=sd, shape=lags)
sigma = pm.HalfNormal('sigma', sigma=1.0)
likelihood = pm.AR('likelihood', beta, sigma=sigma, observed=delta)
trace = pm.sample(1000, progressbar=True, chains=4, cores=1)
plt.plot(delta,alpha=.5)
plt.plot(trace['likelihood'][::100].T, 'r', alpha=.01)
It outputs this error:
Traceback (most recent call last):
File "<ipython-input-10-819006e3a186>", line 1, in <module>
trace[likelihood]
File "/usr/local/lib/python3.6/dist-packages/pymc3/backends/base.py", line 332, in __getitem__
raise KeyError("Unknown variable %s" % var)
KeyError: 'Unknown variable likelihood'
Best wishes,
Gregory
likelihood is an observed variable, and therefore it is NOT sampled during the inference run. It is intended and desired behaviour.
likelihood is defined as a variable to indicate PyMC how to calculate the likelihood, that is, evaluate the probability of the defined distribution AR(beta, sd=sigma) at the observed values, delta in this case. ll_alias is therefore not useful at all to evaluate the fit.
If you pay attention to the plot, you’ll see that all ll_alias samples are identical to delta:
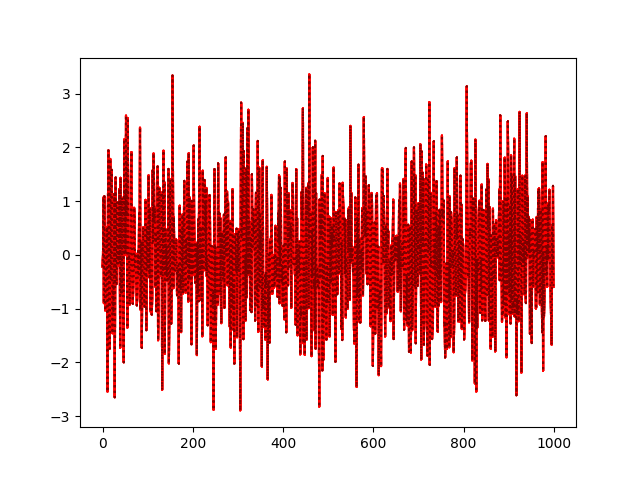
To examine how well does the fitted model describe the observed data, you need posterior predictive samples. If the method has random method implemented (which is not the case for AR distribution), you’ll get the proper samples too examine the fit. See the example notebook on posterior predictive checks and PyMC resources for more details and examples.
@OriolAbril thanks for catching that - sloppy error on my part.