Hi everyone. I have been experimenting with developing models that have a relatively expensive black-box likelihood function (a class of “generalized” drift diffusion models). When trying to sample larger, multilevel models, I have noticed that sampling slows down incredibly as the model in question becomes more “multilevel” (e.g. more levels in a factor and/or more factors that we want to model at once). With sufficiently large models, the computation time is too long to make experimentation at all practical (e.g. easily tens or even hundreds of hours for 1000 samples). I believe I’ve found at one significant culprit and wanted to raise it here.
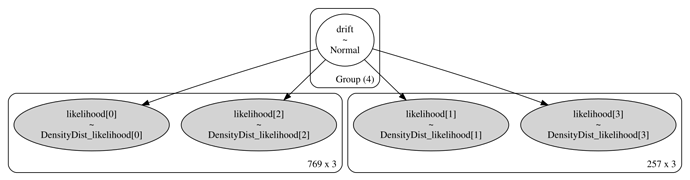
For illustration purposes, suppose we have N observations; and in the data, suppose there is one factor (data column) that has M unique levels. Finally suppose that we want to estimate a model parameter \beta_m for each of the M levels; \beta \in \mathbb{R}^M. In effect, this means that the model will have M likelihood nodes, with the mth likelihood node corresponding to the subset of data observations where the data factor is equal to the mth factor level. Each likelihood node contributes an additive term to the pymc model’s logp().
What I have noticed is that the Slice sampler seems to compute the entire model logp every time it needs a logp calculation, despite the fact that it is a univariate sampling method (only modifying one model parameter at a time, keeping all other parameters fixed). It seems to me that when the sampler is working on \beta_m, it only really needs to be evaluating the likelihood logp from the mth likelihood node—all inputs to the other likelihood nodes are remaining fixed. With a speedy pymc model, this doesn’t seem to be much of an issue. But with an expensive likelihood function, the computational cost compounds noticeably. It is worth noting that the algorithm of the slice sampler makes this particularly pronounced, because it may compute logp() many times for for every univariate proposal.
I may be wrong, but I believe that this point can be restated succinctly using the concept of the Markov blanket: although the univariate sampler working on \beta_m only needs to calculate logps via a forward-pass through the Markov blanket of \beta_m, it calculates the logp of the entire graphical model. In some cases this can be highly redundant.
-
Does this sound correct?
- I have had to learn a good deal just to dig through the codebase and try to track the computation graph that the sampler is calling. I am still learning and could definitely have misunderstood something
 .
.
- I have had to learn a good deal just to dig through the codebase and try to track the computation graph that the sampler is calling. I am still learning and could definitely have misunderstood something
-
Is it feasible to modify the pymc slice sampler to only perform the logp calculation using the relevant subset of the entire model graph? Feasible in terms of: statistically, is it sound; and practically, is it easy enough to do in pymc/aesara.
-
(More to the specific use case.) Does anyone have any other suggestions for me?
- Currently, I have hacked together a pretty sloppy caching mechanism that caches the logp outputs from each logp node, and uses the cached value if a likelihood node is given a parameter set identical to one that it has already seen. This has helped significantly, but has its limitations (cache isn’t shared across chains, introduces overhead, cache size is another tuneable parameter now).
- In very quick-and-dirty tests with smaller models, I haven’t found any other step methods in pymc that seem more promising for sampling these models compared to the slice sampler. But I would be open to suggestions if this particular case (costly likelihood functions, many parameters, but as far as I can tell, not too “pathological” posterior geometries) seems better-suited to some other tool in the pymc toolbox. I am wondering if it may be fruitful to spend some time experimenting with using emcee as a sampler for my models in pymc, since they can at least be massively parallelized — but obviously that’s a pain because I love the pymc models as they are, and know that emcee may not be well-suited to high-dimensional models.
Sorry for the chunky post. I do try to keep it precise… Anyways, thanks as ever!