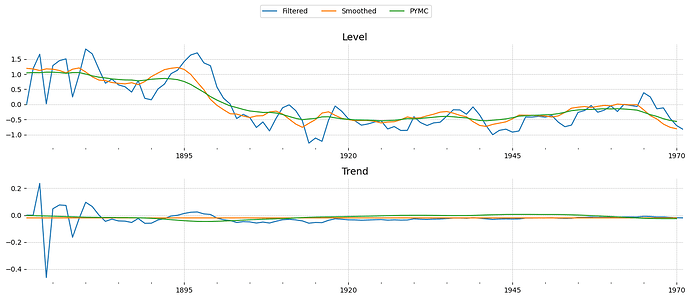
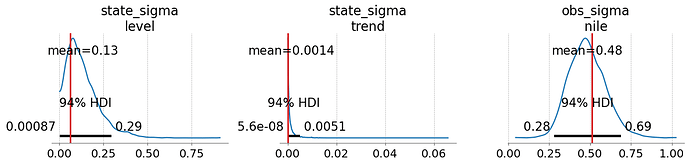
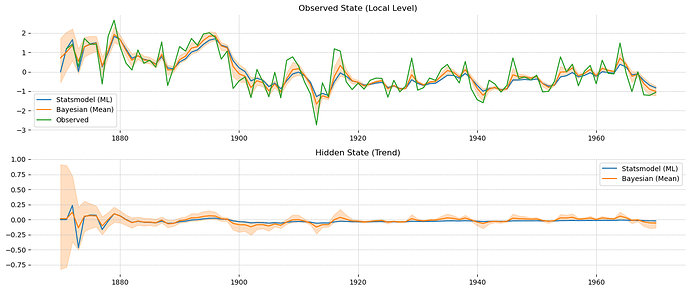
I thought a different angle might be to just implement a Kalman filter in Aesara and use it with pm.Potential to get the model’s likelihood.
I am definitely missing something when it comes to defining an Op, because the shapes are not working out at all. Here’s what I have for the filter:
class KalmanLikelihood(at.Op):
itypes = [at.dmatrix,
at.dmatrix,
at.dmatrix,
at.dmatrix,
at.dmatrix,
at.dmatrix,
at.dmatrix]
otypes = [at.dmatrix, at.dmatrix, at.dscalar]
def __init__(self, data):
self.data = data
self.kalman_filter = self._build_kalman_filter()
def _build_kalman_filter(self):
"""
Construct the computation graph for the Kalman filter
"""
a0 = at.matrix('states', dtype='float32')
P0 = at.matrix('covariance_matrices', dtype='float32')
Q = at.matrix('Q', dtype='float32')
H = at.matrix('H', dtype='float32')
T = at.matrix('T', dtype='float32')
R = at.matrix('R', dtype='float32')
Z = at.matrix('Z', dtype='float32')
data = at.matrix('observed_data', dtype='float32')
matrix_det = at.nlinalg.Det()
matrix_inv = at.nlinalg.matrix_inverse
ll = at.scalar('log_likelihood', dtype='float32')
results, updates = aesara.scan(self._kalman_step,
sequences=[data],
outputs_info=[a0, P0, np.zeros(1, dtype='float32')],
non_sequences=[Q, H, T, R, Z])
states, covariances, log_likelihoods = results
# the scan returns everything shifted forward one time-step, this is a janky fix (is there a more correct solution?)
states = at.concatenate([a0[None, :, :], states], axis=0)
covariances = at.concatenate([P0[None, :, :], covariances], axis=0)
# add unnecessary normalizing constant for comparability with statsmodels
log_likelihood = -data.shape[0] * data.shape[1] / 2 * np.log(2 * np.pi) - 0.5 * log_likelihoods[-1]
return aesara.function([data, a0, P0, Q, H, T, R, Z],
(states, covariances, log_likelihood),
allow_input_downcast=True)
@staticmethod
def _kalman_step(y, a, P, ll, Q, H, T, R, Z):
"""
Conjugate update rule for the mean and covariance matrix, with log-likelihood en passant
"""
v = y - Z.dot(a)
F = Z.dot(P).dot(Z.T) + H
F_inv = matrix_inv(F)
a_update = a + P.dot(Z.T).dot(F_inv).dot(v)
P_update = P - P.dot(Z.T).dot(F_inv).dot(Z).dot(P)
a_hat = T.dot(a_update)
P_hat = T.dot(P_update).dot(T.T) + R.dot(Q).dot(R.T)
ll += (at.log(matrix_det(F)) + (v.T).dot(F_inv).dot(v)).ravel()[0]
return a_hat, P_hat, ll
def perform(self, node, inputs, outputs):
states, covariances, loglikelihood = self.kalman_filter(self.data, *inputs)
outputs[0][0] = states
outputs[1][0] = covariances
outputs[2][0] = loglikelihood
This compiles fine and I can compute the filtered states, covariance, and log-likelihood given the state space matrices and data:
a0 = at.matrix('states', dtype='float64')
P0 = at.matrix('covariance_matrices', dtype='float64')
Q = at.matrix('Q', dtype='float64')
H = at.matrix('H', dtype='float64')
T = at.matrix('T', dtype='float64')
R = at.matrix('R', dtype='float64')
Z = at.matrix('Z', dtype='float64')
f = aesara.function([a0, P0, Q, H, T, R, Z],
KalmanLikelihood(nile.values)(a0, P0, Q, H, T, R, Z))
a = np.array([[0.0],
[0.0]])
P = np.array([[1.0, 0.0],
[0.0, 1.0]])
sigma2_measurement = 1.0
sigma2_level = 1.0
sigma2_trend = 1.0
T_mat = np.array([[1.0, 1.0],
[0.0, 1.0]])
R_mat = np.eye(2)
Z_mat = np.array([[1.0, 0.0]])
Q_mat = np.array([[sigma2_level, 0.0],
[0.0, sigma2_trend]])
H_mat = np.array([sigma2_measurement])[:, None]
f(a, P, Q_mat, H_mat, T_mat, R_mat, Z_mat)
This outputs correct answers. So I was quite pleased with myself, until I tried to put it into a PyMC model:
kl = KalmanLikelihood(nile.values)
with pm.Model(coords=coords) as nile_model:
state_sigmas = pm.HalfNormal('state_sigma', sigma=1.0, dims=['states'], dtype='float64')
H = pm.HalfNormal('observation_noise', sigma=1.0, dtype='float64')
x0 = pm.Normal('x0', mu=0.0, sigma=1.0, dims=['states'], dtype='float64')
sd_dist = pm.Exponential.dist(1.0)
chol, _, _ = pm.LKJCholeskyCov('P0', n=2, eta=1.0,
sd_dist=sd_dist,
compute_corr=True,
store_in_trace=False)
chol = chol.astype('float64')
P0 = at.dot(chol, chol.T)
T = at.as_tensor(np.array([[1.0, 1.0],
[0.0, 1.0]]))
R = at.eye(2, dtype='float64')
Z = at.as_tensor_variable(np.array([1.0, 0.0]))
Q = at.set_subtensor(at.eye(2, dtype='float64')[np.diag_indices(2)], state_sigmas)
pm.Potential("likelihood", kl(x0, P0, Q, H, T, R, Z))
I’m sure there are other problems with this model, but I can’t get to them because Aesara complains about shapes:
TypeError Traceback (most recent call last)
Input In [233], in <cell line: 1>()
21 Z = at.as_tensor_variable(np.array([1.0, 0.0]))
22 Q = at.set_subtensor(at.eye(2, dtype='float64')[np.diag_indices(2)], state_sigmas)
---> 24 pm.Potential("likelihood", kl(x0, P0, Q, H, T, R, Z))
File ~\miniconda3\envs\pymc-dev-py38\lib\site-packages\aesara\graph\op.py:294, in Op.__call__(self, *inputs, **kwargs)
252 r"""Construct an `Apply` node using :meth:`Op.make_node` and return its outputs.
253
254 This method is just a wrapper around :meth:`Op.make_node`.
(...)
291
292 """
293 return_list = kwargs.pop("return_list", False)
--> 294 node = self.make_node(*inputs, **kwargs)
296 if config.compute_test_value != "off":
297 compute_test_value(node)
File ~\miniconda3\envs\pymc-dev-py38\lib\site-packages\aesara\graph\op.py:238, in Op.make_node(self, *inputs)
234 raise ValueError(
235 f"We expected {len(self.itypes)} inputs but got {len(inputs)}."
236 )
237 if not all(it.in_same_class(inp.type) for inp, it in zip(inputs, self.itypes)):
--> 238 raise TypeError(
239 f"Invalid input types for Op {self}:\n"
240 + "\n".join(
241 f"Input {i}/{len(inputs)}: Expected {inp}, got {out}"
242 for i, (inp, out) in enumerate(
243 zip(self.itypes, (inp.type for inp in inputs)),
244 start=1,
245 )
246 if inp != out
247 )
248 )
249 return Apply(self, inputs, [o() for o in self.otypes])
TypeError: Invalid input types for Op KalmanLikelihood:
Input 1/7: Expected TensorType(float64, (None, None)), got TensorType(float64, (None,))
Input 4/7: Expected TensorType(float64, (None, None)), got TensorType(float64, ())
Input 5/7: Expected TensorType(float64, (None, None)), got TensorType(float64, (2, 2))
Input 7/7: Expected TensorType(float64, (None, None)), got TensorType(float64, (2,))
Nothing I do with reshaping makes any difference. For example, wrapping x0 in at.atleast_2d just changes the error to Input 1/7: Expected TensorType(float64, (None, None)), got TensorType(float64, (None, 1)). I also tried using .reshape , fancy indexing [None, :] to add dimensions, and wrapping variables in at.as_tensor_variable(x, ndims=2), but nothing satisfies the (None, None) requirement. I clearly have a fundamental misunderstanding about something here, hopefully someone can help me spot it.
Two other asides. The docstrings for the at.d{scalar/vector/matrix/tensor} family of functions say you can pass dtype and shape arguments, but actually you cannot (I also tried this).
It strikes me as odd that I am compiling the Kalman filter function inside the Op then compiling the op a second time later. I did this because I was just blindly following the black-box tutorial in the docs that says an Op should do a symbolic computation. I imagine some of my problems are connected to this, but I haven’t figured out the clean way to do it. I imagine this way also makes it needlessly difficult to get access to the gradients?