Hi all,
I know there is some great work going on around this thanks to @drbenvincent and others; but I am posting here to see if I can get some more advice and intuition about the approach.
I’ve been working through the examples @drbenvincent’s great notebooks on the ordinal data analysis (GitHub - drbenvincent/ordinal-regression: private development repo for learning ordinal regression topics) try and get a handle on things, but I am still very confused.
I’m looking to use the same approach Kruschke uses in DBDA2 to estimate the latent mean and scale of an ordinal set of data to do some inferences on that. I’ve been using OrderedProbit and have been trying to use the same approach Kruschke uses by pinning the left and right extreme cutpoints, and estimating the mean, scale, and remaining cutpoints. Like the notebooks, I’ve been using the example dataset from DBDA2, OrdinalProbitData-1grp-1.csv, which is stated in the book as having a latent mean of 1 and scale of 2.5.
I’ve been using the following parameterisation (forgive hard-coding of values) which results in relatively sensible estimates (but not of the mean), but with severe divergences, and requires starting values to not crash:
df = pd.read_csv('OrdinalProbitData-1grp-1.csv')
with pm.Model() as model2:
eta = pm.Normal('eta', mu=3, sigma=2)
sigma = pm.HalfNormal('sigma', 1)
cutpoints = at.concatenate([
[0.5],
pm.Uniform('cut_unknown', lower=1, upper=5, shape=4),
[5.5]
])
pm.OrderedProbit('y' cutpoints=cutpoints, eta=eta, sigma=sigma,
compute_p=False, observed=df.Y.values-1)
trace = pm.sample(tune=5000, initvals={'cut_unknown': [1.5, 2.5, 3.5, 4.5]})
az.plot_trace(trace, figsize=(15, 10))
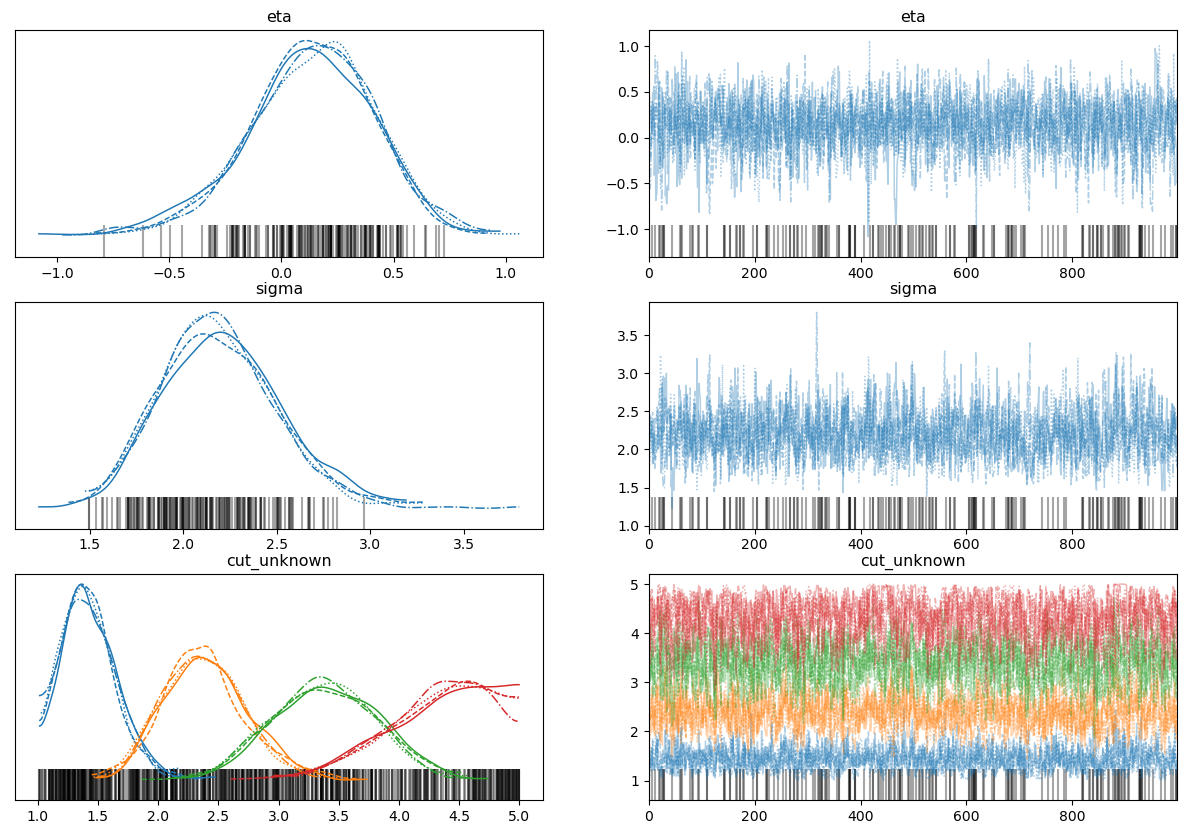
Divergences aside, these look reasonable for the cutpoints and sigma, but not really for the mean (eta). The posterior predictive also looks very reasonable here. I know the cutpoints need to be ordered; I’ve tried transforming the Uniform prior in the code above using distributions.transforms.ordered and that actually increases the number of divergences!
Using the constrainedUniform implementation from @drbenvincent’s notebooks, the divergences disappear as stated, but now the estimates of the mean and scale are very different from 1 and 2.5, and the cutpoints are no longer on the scale of the responses:
def constrainedUniform(N, min=0, max=1):
return pm.Deterministic('theta',
at.concatenate([
np.ones(1)*min,
at.extra_ops.cumsum(pm.Dirichlet("theta_unknown",
a=np.ones(N - 2))) * (min+(max-min))
]), dims='cutpoints')
coords = {'cutpoints': np.arange(6)}
with pm.Model(coords=coords) as model2:
eta = pm.Normal("eta", mu=3, sigma=2)
sigma = pm.HalfNormal('sigma', 1)
cutpoints = constrainedUniform(7)
pm.OrderedProbit("y", cutpoints=cutpoints, eta=eta, sigma=sigma,
compute_p=False, observed=df.Y.values-1)
trace = pm.sample(tune=5000)
az.plot_trace(trace)
I’m probably missing something fundamental here about OrderedProbit, but I’m not sure why having the cutpoints on their cumulative probability scale helps so much, or why the eta and sigma parameters never seem to quite reflect the values stated in DBDA2.
Any pointers are massively appreciated from this struggling Bayesian ![]()