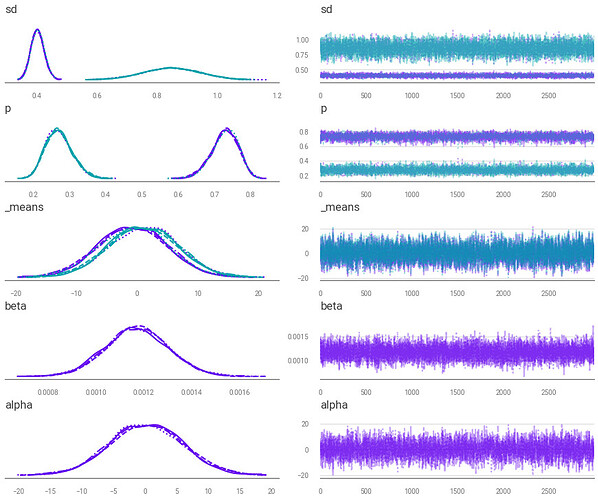
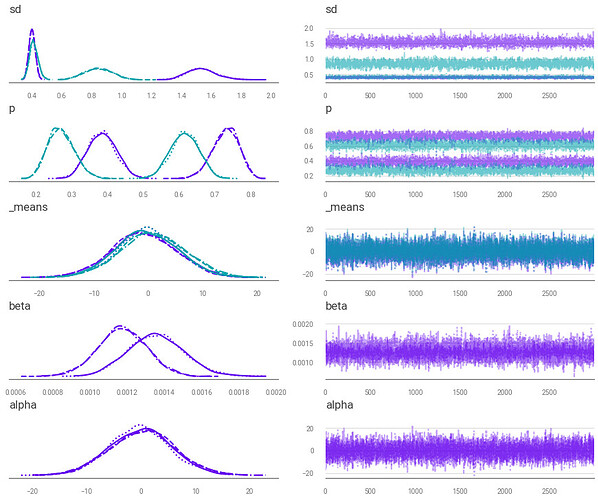
Similar to how a gaussian distribution is fit to a trendline in Bayesian linear regression, I’m attempting to fit a gaussian mixture to a trend line.
I’m having some issues with convergence specifically on the mixture parameters.
As expected for me in a mixture model, I initially get what appears to be some label switching.
I attempt to break symmetry via pm.distributions.transforms.ordered on the means (the line commented out on the code) but this seems to add new peaks to the trace plot.
Why is pm.distributions.transforms.ordered causing these new peaks to appear?
import numpy as np
import pymc3 as pm
import theano.tensor as tt
data = ... # pandas DataFrame containing data
model_usage_data = data[["scaled_value"]]
model_day = model_usage_data.index.to_numpy().reshape(-1, 1)
coords = {"day": model_usage_data.index}
groups = 2
with pm.Model(coords=coords) as model:
alpha = pm.Normal("alpha", mu=0, sd=10)
beta = pm.Normal("beta", mu=0, sd=1)
day_data = pm.Data("day_data", model_day)
broadcast_day = tt.concatenate([day_data, day_data], axis=1)
trend = pm.Deterministic("trend", alpha + beta * broadcast_day)
_means = pm.Normal(
"_means",
mu=[[0, 0.1]],
sd=10,
shape=(1, groups),
# Will be toggling this line
# transform=pm.distributions.transforms.ordered,
testval=np.array([[0, 0.2]]),
)
means = pm.Deterministic("means", _means + trend)
p = pm.Dirichlet("p", a=np.ones(groups))
sds = pm.HalfNormal("sd", sd=10, shape=groups)
pm.NormalMixture("y", w=p, mu=means, sd=sds, observed=model_usage_data)
trace = pm.sample(
draws=draws,
tune=tune,
target_accept=0.90,
max_treedepth=15,
return_inferencedata=False,
)