Those are great resources, thank you. Read through a lot of that and here’s how I proceeded. My first attempt was to do what you suggested, I tried target_accept of 0.9, 0.95 and 0.99. At all of those values, I would non-deterministically get rid of divergences on some runs, but other runs would have between 1-12 divergences. At a value of 0.995, it completely got rid of the divergences. I wanted to explore other options, and I’m curious if this hybrid approach has any merits. I’m also wondering if a target_accept of 0.995 has any negative downsides that might prevent you from wanting to use that value(high auto-correlation, slow convergence?). It also seems very wrong to rely on a model that sometimes will have divergences and sometimes will not.
With a target_accept of 0.95:
with pm.Model() as model:
a = pm.Normal('a', 0., 1.)
sigma = pm.Exponential('sigma', 1.)
a_cluster = pm.Normal('a_cluster', mu=a, sigma=sigma, shape=2)
p = pm.math.invlogit(a_cluster[[0, 1]])
pm.Binomial('obs', p=p, n=[6, 125], observed=[1, 110])
traces = pm.sample(2000, cores=4, target_accept=0.95)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [a_cluster, sigma, a]
Sampling 4 chains, 11 divergences: 100%|██████████| 10000/10000 [00:03<00:00, 3070.63draws/s]
There were 4 divergences after tuning. Increase `target_accept` or reparameterize.
There was 1 divergence after tuning. Increase `target_accept` or reparameterize.
There were 5 divergences after tuning. Increase `target_accept` or reparameterize.
There was 1 divergence after tuning. Increase `target_accept` or reparameterize.
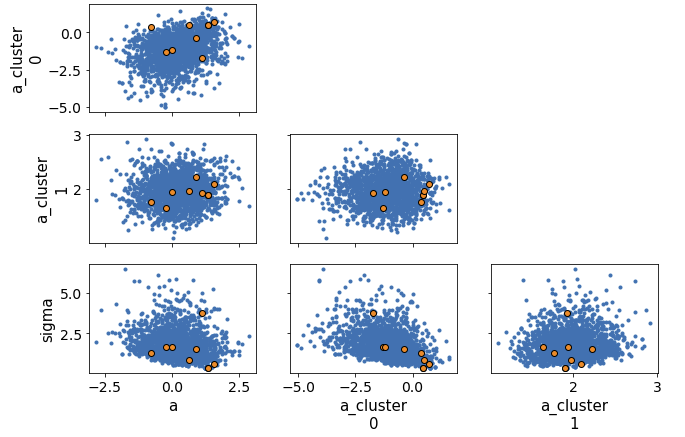
Looking at the pairplot of divergences:
pm.pairplot(traces, divergences=True)
It appears that more divergences are cluster in the corner. I looked at the pairplot of sigma and individual group mean.
x = pd.Series(traces['a_cluster'][:, 0], name='a_cluster')
y = pd.Series(traces['sigma'], name='sigma')
sns.jointplot(x, y, ylim=(0, 2));
This shows a “funnel” shape, which makes sense because as the group sigma gets smaller, it constrains the possible values of the group mean. The downside of this is that it can make it hard for the sampler to efficiently explore the space of lower sigmas. One of the suggestions you linked to talked about using an off-centered reparameterization, so I changed my code to:
with pm.Model() as model:
a = pm.Normal('a', 0., 1.) # group mean
sigma = pm.Exponential('sigma', 1.) # determines amount of shrinkage
a_offset = pm.Normal('a_offset', mu=0, sd=1, shape=2)
a_cluster = pm.Deterministic('a_cluster', a+a_offset*sigma)
p = pm.math.invlogit(a_cluster[[0, 1]])
pm.Binomial('obs', p=p, n=[6, 125], observed=[1, 110])
traces = pm.sample(2000, cores=4, target_accept=0.95)
Which significantly reduced the number of divergences(on some runs I would still get 1 or 2). A target_accept of 0.99 completely removed divergences with this parameterization. A pair plot of the offset and sigma show they’re uncorrelated which (I think) makes it easier for the sampler to explore the space:
x = pd.Series(traces['a_offset'][:, 0], name='a_cluster')
y = pd.Series(traces['sigma'], name='sigma')
sns.jointplot(x, y, ylim=(0, 2));

 ). And these models are the topic of the
). And these models are the topic of the 
 . Thanks for you help!
. Thanks for you help!