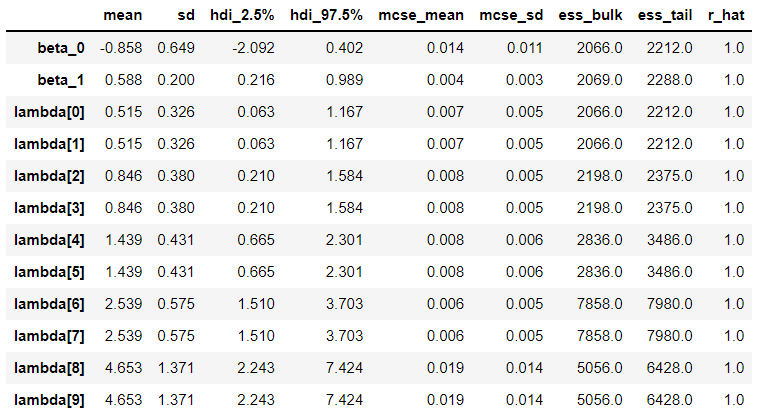
Why does my Deterministic variable (lambda in this case) get split into multiple components in the summary? I see that it’s the same number as the length of my data, but I don’t know where the connection is.
Here is the code:
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pymc3 as pm
X = np.array((0., 0., 1., 1., 2., 2., 3., 3., 4., 4.))
y = np.array((0., 0., 1., 1., 2., 2., 3., 3., 4., 4.))
with pm.Model() as m:
# priors
beta_0 = pm.Normal("beta_0", mu=0, sigma=1000)
beta_1 = pm.Normal("beta_1", mu=0, sigma=1000)
lambda_ = pm.Deterministic("lambda", pm.math.exp(beta_0 + beta_1 * X))
# likelihood
y_pred = pm.Poisson("y_pred", lambda_, observed=y)
# start sampling
trace = pm.sample(
3000, # samples
chains=4,
tune=1000,
init="jitter+adapt_diag",
random_seed=1,
return_inferencedata=True,
)
az.summary(trace, hdi_prob=0.95)
Here is the summary output: