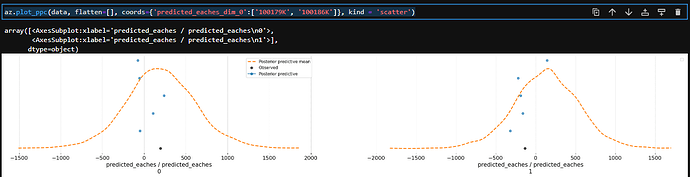
Thank you. I changed to the following:
obs_item = idata.posterior["item"][idata.constant_data["ic_to_item_map_dim_0"]]
data = idata.assign_coords(obs_id=obs_item, groups="observed_vars")
az.plot_ppc(data, coords={'ic_to_item_map_dim_0': ['110656K']}, flatten=[], kind='cumulative')
This produces 40 graphs for one item where I’m aiming for one graph per item. In the code above, I just pull one item.
Maybe this has to do with my model?
Here is my model for reference:
coords = {'business_line':business_lines,
'category':categories,
'subcategory':subcats,
'ic':ic,
'item':items,
'time':times,
'location':locations,
'promo': promo,
'giftset':giftset,
'free_fin':free_fin,
'dc_discount':dc_discount,
# 'month':month,
'cannibalization':cannibalization,
# 'yearly_components':[f'yearly_{f}_{i+1}' for f in ['cos', 'sin'] for i in range(x_fourier.shape[1] // 2)],
# 'weekly_components':[f'weekly_{f}_{i+1}' for f in ['cos', 'sin'] for i in range(x_fourier_week.shape[1] // 2)]
}
#TRAINING THE MODEL STARTS HERE
print("fitting model...")
with pm.Model(coords=coords) as constant_model:
#Data that does not change
cat_to_bl_map = pm.Data('cat_to_bl_map', cat_to_bl_idx, mutable=False)
subcat_to_cat_map = pm.Data('subcat_to_cat_map', subcat_to_cat_idx, mutable=False)
ic_to_subcat_map = pm.Data('ic_to_subcat_map', ic_to_subcat_idx, mutable=False)
ic_to_item_map = pm.Data('ic_to_item_map', ic_to_item_idx, mutable = False)
#Data that does change
pm_loc_idx = pm.Data('loc_idx', location_idx, mutable = True)
pm_item_idx = pm.Data('item_idx', item_idx, mutable=True)
pm_time_idx = pm.Data('time_idx', time_idx, mutable=True)
observed_eaches = pm.Data('observed_eaches', df.residual, mutable=True)
promo_ = pm.Data('promotion', promo_idx, mutable = True)
cann_ = pm.Data('cannibalization', cann_idx, mutable = True)
dc_discount_ = pm.Data('dc_discount', dc_idx, mutable = True)
free_fin_ = pm.Data('free_fin', free_fin_idx, mutable = True)
pvbv_ = pm.Data('pvbv', promo_pvbv_idx, mutable = True)
giftset_ = pm.Data('giftset', giftset_idx, mutable = True)
month_2_ = pm.Data('month_2', month_2_idx, mutable = True)
month_3_ = pm.Data('month_3', month_3_idx ,mutable = True)
month_4_ = pm.Data('month_4', month_4_idx ,mutable = True)
month_5_ = pm.Data('month_5', month_5_idx ,mutable = True)
month_6_ = pm.Data('month_6', month_6_idx ,mutable = True)
month_7_ = pm.Data('month_7', month_7_idx ,mutable = True)
month_8_ = pm.Data('month_8', month_8_idx ,mutable = True)
month_9_ = pm.Data('month_9', month_9_idx ,mutable = True)
month_10_ = pm.Data('month_10', month_10_idx,mutable = True)
month_11_ = pm.Data('month_11', month_11_idx ,mutable = True)
month_12_ = pm.Data('month_12', month_12_idx ,mutable = True)
# dcost_local_change_ = pm.Data('dcost_local', price_idx, mutable = True)
price_change_before_ = pm.Data('price_change_before', price_before_idx, mutable = True)
price_change_on_ = pm.Data('price_change_on', price_before_idx, mutable = True)
loc_intercept = pm.Normal('loc_intercept', mu = 0, sigma = 1, dims = ['location'])
loc_bl = utility_functions.make_next_level_hierarchy_variable(name='loc_bl', mu=loc_intercept, alpha=2, beta=1, dims=['business_line', 'location'])
loc_cat = utility_functions.make_next_level_hierarchy_variable(name='loc_cat', mu=loc_bl[cat_to_bl_map], alpha=2, beta=1, dims=['category', 'location'])
loc_subcat = utility_functions.make_next_level_hierarchy_variable(name='loc_subcat', mu=loc_cat[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory', 'location'])
loc_ic = utility_functions.make_next_level_hierarchy_variable(name='loc_ic', mu=loc_subcat[ic_to_subcat_map], alpha=2, beta=1, dims=['ic', 'location'])
loc_item = utility_functions.make_next_level_hierarchy_variable(name='loc_item', mu=loc_ic[ic_to_item_map], alpha=2, beta=1, dims=['item', 'location'])
mu_cann = pm.Normal('mu_cann', mu = 0, sigma = 1)
bl_cann = utility_functions.make_next_level_hierarchy_variable(name='bl_cann', mu=mu_cann, alpha=2, beta=1, dims=['business_line'])
cat_cann = utility_functions.make_next_level_hierarchy_variable(name='cat_cann', mu=bl_cann[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_cann = utility_functions.make_next_level_hierarchy_variable(name='subcat_cann', mu=cat_cann[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_cann = utility_functions.make_next_level_hierarchy_variable(name='ic_cann', mu=subcat_cann[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_cann = utility_functions.make_next_level_hierarchy_variable(name='item_cann', mu=ic_cann[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_dc_discount = pm.Normal('mu_dc_discount', mu = 0, sigma = 1)
bl_dc_discount = utility_functions.make_next_level_hierarchy_variable(name='bl_dc_discount', mu=mu_dc_discount, alpha=2, beta=1, dims=['business_line'])
cat_dc_discount = utility_functions.make_next_level_hierarchy_variable(name='cat_dc_discount', mu=bl_dc_discount[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_dc_discount = utility_functions.make_next_level_hierarchy_variable(name='subcat_dc_discount', mu=cat_dc_discount[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_dc_discount = utility_functions.make_next_level_hierarchy_variable(name='ic_dc_discount', mu=subcat_dc_discount[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_dc_discount = utility_functions.make_next_level_hierarchy_variable(name='item_dc_discount', mu=ic_dc_discount[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_free_fin = pm.Normal('mu_free_fin', mu = 0, sigma = 1)
bl_free_fin = utility_functions.make_next_level_hierarchy_variable(name='bl_free_fin', mu=mu_free_fin, alpha=2, beta=1, dims=['business_line'])
cat_free_fin = utility_functions.make_next_level_hierarchy_variable(name='cat_free_fin', mu=bl_free_fin[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_free_fin = utility_functions.make_next_level_hierarchy_variable(name='subcat_free_fin', mu=cat_free_fin[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_free_fin = utility_functions.make_next_level_hierarchy_variable(name='ic_free_fin', mu=subcat_free_fin[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_free_fin = utility_functions.make_next_level_hierarchy_variable(name='item_free_fin', mu=ic_free_fin[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_pvbv = pm.Normal('mu_pvbv', mu = 0, sigma = 1)
bl_pvbv = utility_functions.make_next_level_hierarchy_variable(name='bl_pvbv', mu=mu_pvbv, alpha=2, beta=1, dims=['business_line'])
cat_pvbv = utility_functions.make_next_level_hierarchy_variable(name='cat_pvbv', mu=bl_pvbv[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_pvbv = utility_functions.make_next_level_hierarchy_variable(name='subcat_pvbv', mu=cat_pvbv[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_pvbv = utility_functions.make_next_level_hierarchy_variable(name='ic_pvbv', mu=subcat_pvbv[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_pvbv = utility_functions.make_next_level_hierarchy_variable(name='item_pvbv', mu=ic_pvbv[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_giftset = pm.Normal('mu_giftset', mu = 0, sigma = 1)
bl_giftset = utility_functions.make_next_level_hierarchy_variable(name='bl_giftset', mu=mu_giftset, alpha=2, beta=1, dims=['business_line'])
cat_giftset = utility_functions.make_next_level_hierarchy_variable(name='cat_giftset', mu=bl_giftset[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_giftset = utility_functions.make_next_level_hierarchy_variable(name='subcat_giftset', mu=cat_giftset[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_giftset = utility_functions.make_next_level_hierarchy_variable(name='ic_giftset', mu=subcat_giftset[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_giftset = utility_functions.make_next_level_hierarchy_variable(name='item_giftset', mu=ic_giftset[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month2 = pm.Normal('mu_month2', mu = 0, sigma = 1)
bl_month2 = utility_functions.make_next_level_hierarchy_variable(name='bl_month2', mu=mu_month2, alpha=2, beta=1, dims=['business_line'])
cat_month2 = utility_functions.make_next_level_hierarchy_variable(name='cat_month2', mu=bl_month2[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month2 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month2', mu=cat_month2[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month2 = utility_functions.make_next_level_hierarchy_variable(name='ic_month2', mu=subcat_month2[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month2 = utility_functions.make_next_level_hierarchy_variable(name='item_month2', mu=ic_month2[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month3 = pm.Normal('mu_month3', mu = 0, sigma = 1)
bl_month3 = utility_functions.make_next_level_hierarchy_variable(name='bl_month3', mu=mu_month3, alpha=2, beta=1, dims=['business_line'])
cat_month3 = utility_functions.make_next_level_hierarchy_variable(name='cat_month3', mu=bl_month3[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month3 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month3', mu=cat_month3[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month3 = utility_functions.make_next_level_hierarchy_variable(name='ic_month3', mu=subcat_month3[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month3 = utility_functions.make_next_level_hierarchy_variable(name='item_month3', mu=ic_month3[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month4 = pm.Normal('mu_month4', mu = 0, sigma = 1)
bl_month4 = utility_functions.make_next_level_hierarchy_variable(name='bl_month4', mu=mu_month4, alpha=2, beta=1, dims=['business_line'])
cat_month4 = utility_functions.make_next_level_hierarchy_variable(name='cat_month4', mu=bl_month4[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month4 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month4', mu=cat_month4[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month4 = utility_functions.make_next_level_hierarchy_variable(name='ic_month4', mu=subcat_month4[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month4 = utility_functions.make_next_level_hierarchy_variable(name='item_month4', mu=ic_month4[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month5 = pm.Normal('mu_month5', mu = 0, sigma = 1)
bl_month5 = utility_functions.make_next_level_hierarchy_variable(name='bl_month5', mu=mu_month5, alpha=2, beta=1, dims=['business_line'])
cat_month5 = utility_functions.make_next_level_hierarchy_variable(name='cat_month5', mu=bl_month5[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month5 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month5', mu=cat_month5[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month5 = utility_functions.make_next_level_hierarchy_variable(name='ic_month5', mu=subcat_month5[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month5 = utility_functions.make_next_level_hierarchy_variable(name='item_month5', mu=ic_month5[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month6 = pm.Normal('mu_month6', mu = 0, sigma = 1)
bl_month6 = utility_functions.make_next_level_hierarchy_variable(name='bl_month6', mu=mu_month6, alpha=2, beta=1, dims=['business_line'])
cat_month6 = utility_functions.make_next_level_hierarchy_variable(name='cat_month6', mu=bl_month6[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month6 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month6', mu=cat_month6[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month6 = utility_functions.make_next_level_hierarchy_variable(name='ic_month6', mu=subcat_month6[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month6 = utility_functions.make_next_level_hierarchy_variable(name='item_month6', mu=ic_month6[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month7 = pm.Normal('mu_month7', mu = 0, sigma = 1)
bl_month7 = utility_functions.make_next_level_hierarchy_variable(name='bl_month7', mu=mu_month7, alpha=2, beta=1, dims=['business_line'])
cat_month7 = utility_functions.make_next_level_hierarchy_variable(name='cat_month7', mu=bl_month7[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month7 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month7', mu=cat_month7[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month7 = utility_functions.make_next_level_hierarchy_variable(name='ic_month7', mu=subcat_month7[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month7 = utility_functions.make_next_level_hierarchy_variable(name='item_month7', mu=ic_month7[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month8 = pm.Normal('mu_month8', mu = 0, sigma = 1)
bl_month8 = utility_functions.make_next_level_hierarchy_variable(name='bl_month8', mu=mu_month8, alpha=2, beta=1, dims=['business_line'])
cat_month8 = utility_functions.make_next_level_hierarchy_variable(name='cat_month8', mu=bl_month8[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month8 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month8', mu=cat_month8[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month8 = utility_functions.make_next_level_hierarchy_variable(name='ic_month8', mu=subcat_month8[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month8 = utility_functions.make_next_level_hierarchy_variable(name='item_month8', mu=ic_month8[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month9 = pm.Normal('mu_month9', mu = 0, sigma = 5)
bl_month9 = utility_functions.make_next_level_hierarchy_variable(name='bl_month9', mu=mu_month9, alpha=2, beta=1, dims=['business_line'])
cat_month9 = utility_functions.make_next_level_hierarchy_variable(name='cat_month9', mu=bl_month9[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month9 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month9', mu=cat_month9[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month9 = utility_functions.make_next_level_hierarchy_variable(name='ic_month9', mu=subcat_month9[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month9 = utility_functions.make_next_level_hierarchy_variable(name='item_month9', mu=ic_month9[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month10 = pm.Normal('mu_month10', mu = 0, sigma = 1)
bl_month10 = utility_functions.make_next_level_hierarchy_variable(name='bl_month10', mu=mu_month10, alpha=2, beta=1, dims=['business_line'])
cat_month10 = utility_functions.make_next_level_hierarchy_variable(name='cat_month10', mu=bl_month10[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month10 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month10', mu=cat_month10[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month10 = utility_functions.make_next_level_hierarchy_variable(name='ic_month10', mu=subcat_month10[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month10 = utility_functions.make_next_level_hierarchy_variable(name='item_month10', mu=ic_month10[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month11 = pm.Normal('mu_month11', mu = 0, sigma = 1)
bl_month11 = utility_functions.make_next_level_hierarchy_variable(name='bl_month11', mu=mu_month11, alpha=2, beta=1, dims=['business_line'])
cat_month11 = utility_functions.make_next_level_hierarchy_variable(name='cat_month11', mu=bl_month11[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month11 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month11', mu=cat_month11[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month11 = utility_functions.make_next_level_hierarchy_variable(name='ic_month11', mu=subcat_month11[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month11 = utility_functions.make_next_level_hierarchy_variable(name='item_month11', mu=ic_month11[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_month12 = pm.Normal('mu_month12', mu = 0, sigma = 1)
bl_month12 = utility_functions.make_next_level_hierarchy_variable(name='bl_month12', mu=mu_month12, alpha=2, beta=1, dims=['business_line'])
cat_month12 = utility_functions.make_next_level_hierarchy_variable(name='cat_month12', mu=bl_month12[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_month12 = utility_functions.make_next_level_hierarchy_variable(name='subcat_month12', mu=cat_month12[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_month12 = utility_functions.make_next_level_hierarchy_variable(name='ic_month12', mu=subcat_month12[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_month12 = utility_functions.make_next_level_hierarchy_variable(name='item_month12', mu=ic_month12[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_price_change = pm.Normal('mu_price_change', mu = 0, sigma = 1)
bl_price_change = utility_functions.make_next_level_hierarchy_variable(name='bl_price_change', mu=mu_price_change, alpha=2, beta=1, dims=['business_line'])
cat_price_change = utility_functions.make_next_level_hierarchy_variable(name='cat_price_change', mu=bl_price_change[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_price_change = utility_functions.make_next_level_hierarchy_variable(name='subcat_price_change', mu=cat_price_change[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_price_change = utility_functions.make_next_level_hierarchy_variable(name='ic_price_change', mu=subcat_price_change[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_price_change = utility_functions.make_next_level_hierarchy_variable(name='item_price_change', mu=ic_price_change[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu_price_change_after = pm.Normal('mu_price_change_after', mu = 0, sigma = 1)
bl_price_change_after = utility_functions.make_next_level_hierarchy_variable(name='bl_price_change_after', mu=mu_price_change_after, alpha=2, beta=1, dims=['business_line'])
cat_price_change_after = utility_functions.make_next_level_hierarchy_variable(name='cat_price_change_after', mu=bl_price_change_after[cat_to_bl_map], alpha=2, beta=1, dims=['category'])
subcat_price_change_after = utility_functions.make_next_level_hierarchy_variable(name='subcat_price_change_after', mu=cat_price_change_after[subcat_to_cat_map], alpha=2, beta=1, dims=['subcategory'])
ic_price_change_after = utility_functions.make_next_level_hierarchy_variable(name='ic_price_change_after', mu=subcat_price_change_after[ic_to_subcat_map], alpha=2, beta=1, dims=['ic'])
item_price_change_after = utility_functions.make_next_level_hierarchy_variable(name='item_price_change_after', mu=ic_price_change_after[ic_to_item_map], alpha=2, beta=1, dims=['item'])
mu = (loc_item[pm_item_idx, pm_loc_idx] + item_cann[pm_item_idx]*cann_ + item_dc_discount[pm_item_idx]*dc_discount_
+ item_free_fin[pm_item_idx]*free_fin_ + item_pvbv[pm_item_idx]*pvbv_ + item_giftset[pm_item_idx]*giftset_ + item_month2[pm_item_idx]*month_2_
+ item_month2[pm_item_idx]*month_3_ + item_month2[pm_item_idx]*month_4_ + item_month2[pm_item_idx]*month_5_ + item_month2[pm_item_idx]*month_6_
+ item_month2[pm_item_idx]*month_7_ + item_month2[pm_item_idx]*month_8_ + item_month2[pm_item_idx]*month_9_ + item_month2[pm_item_idx]*month_10_
+ item_month2[pm_item_idx]*month_11_ + item_month2[pm_item_idx]*month_12_ + item_price_change[pm_item_idx]*price_change_before_
+ item_price_change_after[pm_item_idx]*price_change_on_)
sigma = pm.HalfNormal('sigma', sigma=100)
eaches = pm.StudentT('predicted_eaches',
mu=mu,
sigma=sigma,
nu=15,
# lower = 0,
observed=observed_eaches)
I’m not sure what is causing so many graphs.