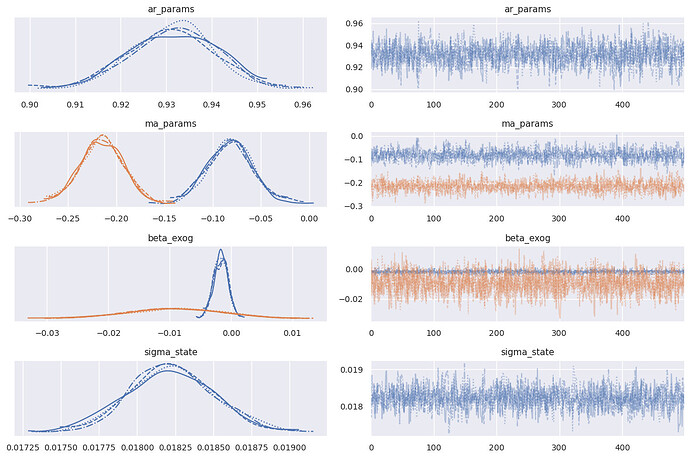
I am trying to model an industrial process in which corn and water are mixed together and cooked in 2 large mixing tanks before flowing out to the next step of the process. There is a density meter on the outflow. The system runs continuously so there is almost always water and corn coming in and the cooked product coming out. Here is a graph of the standardized corn, water, and density:
The x axis is in minutes.
I want to build a model that predicts the density given the corn and water flow rates. More corn and less water makes the density go up but because of mixing there is a time delay and the effect of turning off the grind for a few minutes doesn’t produce an immediate and corresponding drop in the density. The density decreases gradually in response to the corn flow stopping then comes back up gradually after the water flow is reduced. I also have other variables I would like to include such as the temperatures and levels of both tanks and the flow rate out of the entire system which might affect the lag time.
What kind of model can I use for this? I have been experimenting with BayesianVARMAX by trying the notebook here but it takes multiple hours to sample a small subset of the data. I also don’t see a place where I can put in exogenous vs endogenous variables.
Should I use gaussian processes? gaussian random walks? Some other kind of model?