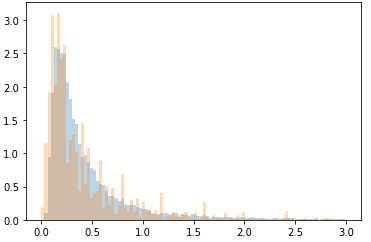
This is a relatively simple question. I’ve had no issues working with generated distributions, or any kind of “toy data”, but now that I have attempted to use some real world data that is a bit more messy I am having issues.
Here is the real data
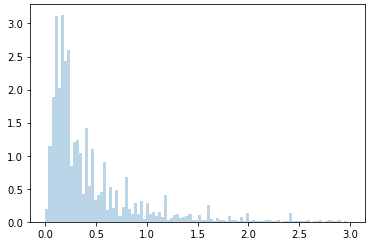
Here is the data I generating using a best fit inverse Gaussian distribution
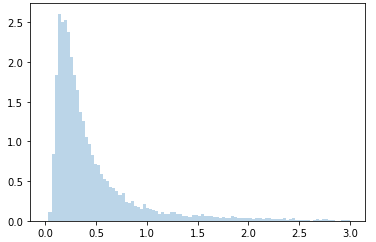
I am successful in imputing missing values from the generated data as follows:
with pm.Model() as model:
initial = np.random.randn(ma.count_masked(test)).astype('float64')
#init = np.abs(np.random.randn(14990,1))
alpha_m=pm.HalfNormal('alpha_m',1,testval=1)
beta_m=pm.HalfNormal('beta_m',1,testval=1)
mu_m=pm.Normal('mu_m',0,1,testval=0)
x = pm.InverseGamma('x',alpha=alpha_m,beta=beta_m,mu=mu_m,observed=data
approx=pm.sample(2000,tune=2000,progressbar=True,chains=1,init='adapt_diag',target_accept=.99,discard_tuned_samples=True)
However, when using the “real world” data set the sampling fails immediately with the "Mass matrix contains zeros on the diagonal. " error. I’ve tried changing the priors, changing the sampling technique, and quite a few other things but I am at a loss. Is there something I am missing in terms of sampling from a messy distribution like this?