Thanks for the help so far with this model!
I am fitting a reinforcement learning model to human behavior in a hierarchical learning task. The model selects actions based on action values (Q-values), which are calculated from trial-by-trial reward prediction errors (RPEs) based on action feedback.
The data can be found here.
import pymc3 as pm
import theano
import theano.tensor as T
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Sampling
n_samples = 100
n_tune = 50
n_cores = 1
n_chains = 1
# Get data
n_seasons, n_TS, n_aliens, n_actions = 3, 3, 4, 3
n_trials, n_subj = 440, 31
alien_initial_Q = 0.12
seasons = T.as_tensor_variable(pd.read_csv("seasons.csv")) # stimulus part 1
aliens = T.as_tensor_variable(pd.read_csv("aliens.csv")) # stimulus part 2
actions = T.as_tensor_variable(pd.read_csv("actions.csv")) # participants' selected actions
rewards = T.as_tensor_variable(pd.read_csv("rewards.csv")) # participants' received rewards
trials, subj = np.meshgrid(range(n_trials), range(n_subj))
trials = T.as_tensor_variable(trials.T)
subj = T.as_tensor_variable(subj.T)
# Fit model
with pm.Model() as model:
# RL parameters: softmax temperature beta; learning rate alpha; Q-value forgetting
beta_mu = pm.Uniform('beta_mu', lower=0, upper=5, testval=1.25)
beta_sd = pm.Uniform('beta_sd', lower=0, upper=5, testval=0.1)
beta_matt = pm.Normal('beta_matt', mu=0, sd=1, shape=n_subj, testval=np.random.choice([-0.1, 0, 0.1], n_subj))
beta = pm.Deterministic('beta', beta_mu + beta_sd * beta_matt)
alpha_mu = pm.Uniform('alpha_mu', lower=0, upper=1, testval=0.15)
alpha_sd = pm.Uniform('alpha_sd', lower=0, upper=1, testval=0.05)
alpha_matt = pm.Normal('alpha_matt', mu=0, sd=1, shape=n_subj, testval=np.random.choice([-1, 0, 1], n_subj))
alpha = pm.Deterministic('alpha', alpha_mu + alpha_sd * alpha_matt)
forget_mu = pm.Uniform('forget_mu', lower=0, upper=1, testval=0.06)
forget_sd = pm.Uniform('forget_sd', lower=0, upper=1, testval=0.03)
forget_matt = pm.Normal('forget_matt', mu=0, sd=1, shape=n_subj, testval=np.random.choice([-1, 0, 1], n_subj))
forget = pm.Deterministic('forget', forget_mu + forget_sd * forget_matt)
# Get the right shapes
betas = T.tile(T.repeat(beta, n_actions), n_trials).reshape([n_trials, n_subj, n_actions]) # Q_sub.shape
forgets = T.repeat(forget, n_TS * n_aliens * n_actions).reshape([n_subj, n_TS, n_aliens, n_actions]) # Q_low for 1 trial
# Initialize Q-values
Q_low0 = alien_initial_Q * T.ones([n_subj, n_TS, n_aliens, n_actions])
# Update Q-values based on stimulus, action, and reward
def update_Qs(season, alien, action, reward, Q_low, alpha, forget, n_subj):
# Loop over trials: take data for all subjects, 1 single trial
# Forget Q-values a little bit in each trial
Q_low_new = (1 - forget) * Q_low + forget * alien_initial_Q * T.ones_like(Q_low)
# Calculate RPE & update Q-values
RPE_low = reward - Q_low_new[T.arange(n_subj), season, alien, action]
Q_low_new = T.set_subtensor(Q_low_new[T.arange(n_subj), season, alien, action],
Q_low_new[T.arange(n_subj), season, alien, action] + alpha * RPE_low)
return Q_low_new
# Get Q-values for the whole task (update Q-values in each trial)
Q_low, _ = theano.scan(fn=update_Qs,
sequences=[seasons, aliens, actions, rewards],
outputs_info=[Q_low0],
non_sequences=[alpha, forgets, n_subj])
Q_low = T.concatenate([[Q_low0], Q_low[:-1]], axis=0) # Add first trial's Q-values, remove last trials Q-values
T.printing.Print("Q_low")(Q_low)
# Subset Q-values according to stimuli and responses in each trial
Q_sub = Q_low[trials, subj, seasons, aliens]
# First step to translate into probabilities: apply exp
p = T.exp(betas * Q_sub)
# Bring p in the correct shape for pm.Categorical: 2-d array of trials
action_wise_p = p.reshape([n_trials * n_subj, n_actions])
action_wise_actions = actions.flatten()
# Select actions (& second step of calculating probabilities: normalize)
actions = pm.Categorical('actions', p=action_wise_p, observed=action_wise_actions)
# Sample the model
trace = pm.sample(n_samples, tune=n_tune, chains=n_chains, cores=n_cores, nuts_kwargs=dict(target_accept=.80))
pm.traceplot(trace)
plt.savefig('trace_plot.png')
model_summary = pm.summary(trace)
pd.DataFrame(model_summary).to_csv('model_summary.csv')
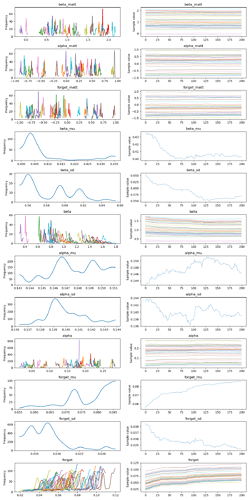
The traceplot looks like this, which seems reasonable given the small number of sampling steps. The values of alpha, beta, and forget make absolute sense for the task:
I don’t have a lot of experience using PyMC3. Does the model and the results look reasonable so far?
What I would like to do next, and where I am running into problems, is adding hierarchy to the RL model. Currently, the model stores the Q-values separately for each season-alien-action compound. What I would like to do instead is to learn the Q-values for each season inside a separate “task set” (TS).
Here is my attempt for modeling (the main difference is within update_Qs):
import pymc3 as pm
import theano
import theano.tensor as T
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Sampling
n_samples = 200
n_tune = 50
n_cores = 1
n_chains = 1
# Get data
n_seasons, n_TS, n_aliens, n_actions = 3, 3, 4, 3
n_trials, n_subj = 440, 31
alien_initial_Q = 0.12
seasons = T.as_tensor_variable(pd.read_csv("seasons.csv"))
aliens = T.as_tensor_variable(pd.read_csv("aliens.csv"))
actions = T.as_tensor_variable(pd.read_csv("actions.csv"))
rewards = T.as_tensor_variable(pd.read_csv("rewards.csv"))
trials, subj = np.meshgrid(range(n_trials), range(n_subj))
trials = T.as_tensor_variable(trials.T)
subj = T.as_tensor_variable(subj.T)
# Fit model
with pm.Model() as model:
# RL parameters: softmax temperature beta; learning rate alpha; Q-value forgetting
beta_mu = pm.Uniform('beta_mu', lower=0, upper=5, testval=1.25)
beta_sd = pm.Uniform('beta_sd', lower=0, upper=5, testval=0.1)
beta_matt = pm.Normal('beta_matt', mu=0, sd=1, shape=n_subj, testval=np.random.choice([-0.1, 0, 0.1], n_subj))
beta = pm.Deterministic('beta', beta_mu + beta_sd * beta_matt)
alpha_mu = pm.Uniform('alpha_mu', lower=0, upper=1, testval=0.15)
alpha_sd = pm.Uniform('alpha_sd', lower=0, upper=1, testval=0.05)
alpha_matt = pm.Normal('alpha_matt', mu=0, sd=1, shape=n_subj, testval=np.random.choice([-1, 0, 1], n_subj))
alpha = pm.Deterministic('alpha', alpha_mu + alpha_sd * alpha_matt)
forget_mu = pm.Uniform('forget_mu', lower=0, upper=1, testval=0.06)
forget_sd = pm.Uniform('forget_sd', lower=0, upper=1, testval=0.03)
forget_matt = pm.Normal('forget_matt', mu=0, sd=1, shape=n_subj, testval=np.random.choice([-1, 0, 1], n_subj))
forget = pm.Deterministic('forget', forget_mu + forget_sd * forget_matt)
beta_high = beta.copy()
alpha_high = alpha.copy()
forget_high = forget.copy()
# Get the right shapes
betas = T.tile(T.repeat(beta, n_actions), n_trials).reshape([n_trials, n_subj, n_actions]) # Q_sub.shape
beta_highs = T.repeat(beta_high, n_TS).reshape([n_subj, n_TS]) # Q_high_sub.shape
forgets = T.repeat(forget, n_TS * n_aliens * n_actions).reshape([n_subj, n_TS, n_aliens, n_actions]) # Q_low for 1 trial
forget_highs = T.repeat(forget_high, n_seasons * n_TS).reshape([n_subj, n_seasons, n_TS]) # Q_high for 1 trial
# Initialize Q-values
Q_low0 = alien_initial_Q * T.ones([n_subj, n_TS, n_aliens, n_actions])
Q_high0 = alien_initial_Q * T.ones([n_subj, n_seasons, n_TS])
# Update Q-values based on stimulus, action, and reward
def update_Qs(season, alien, action, reward, Q_low, Q_high, alpha, alpha_high, beta_high, forget, forget_high,
n_subj):
# Loop over trials: take data for all subjects, 1 single trial
# Select TS
Q_high_sub = Q_high[T.arange(n_subj), season]
p_high = T.exp(beta_high * Q_high_sub)
# TS = season # Use this line instead of the next one to get similar results as for the flat model above
TS = p_high.argmax(axis=1) # Select one TS deterministically
# Forget Q-values a little bit
Q_low_new = (1 - forget) * Q_low + forget * alien_initial_Q * T.ones_like(Q_low)
Q_high_new = (1 - forget_high) * Q_high + forget_high * alien_initial_Q * T.ones_like(Q_high)
# Calculate RPEs & update Q-values
RPE_low = reward - Q_low_new[T.arange(n_subj), TS, alien, action]
Q_low_new = T.set_subtensor(Q_low_new[T.arange(n_subj), TS, alien, action],
Q_low_new[T.arange(n_subj), TS, alien, action] + alpha * RPE_low)
RPE_high = reward - Q_high_new[T.arange(n_subj), season, TS]
Q_high_new = T.set_subtensor(Q_high_new[T.arange(n_subj), season, TS],
Q_high_new[T.arange(n_subj), season, TS] + alpha_high * RPE_high)
return [Q_low_new, Q_high_new]
# Get Q-values for the whole task (update each trial)
[Q_low, Q_high], _ = theano.scan(fn=update_Qs, # hierarchical: TS = p_high.argmax(axis=1); flat: TS = season
sequences=[seasons, aliens, actions, rewards],
outputs_info=[Q_low0, Q_high0],
non_sequences=[alpha, alpha_high, beta_highs, forgets, forget_highs, n_subj])
Q_low = T.concatenate([[Q_low0], Q_low[:-1]], axis=0) # Add first trial's Q-values, remove last trials Q-values
T.printing.Print("Q_low")(Q_low)
T.printing.Print("Q_high")(Q_high)
# Subset Q-values according to stimuli and responses in each trial
Q_sub = Q_low[trials, subj, seasons, aliens]
# First step to translate into probabilities: apply exp
p = T.exp(betas * Q_sub)
# Bring p in the correct shape for pm.Categorical: 2-d array of trials
action_wise_p = p.reshape([n_trials * n_subj, n_actions])
action_wise_actions = actions.flatten()
# Select actions (& second step of calculating probabilities: normalize)
actions = pm.Categorical('actions', p=action_wise_p, observed=action_wise_actions)
# Sample the model
trace = pm.sample(n_samples, tune=n_tune, chains=n_chains, cores=n_cores, nuts_kwargs=dict(target_accept=.80))
pm.traceplot(trace)
plt.savefig('trace_plot.png')
model_summary = pm.summary(trace)
pd.DataFrame(model_summary).to_csv('model_summary.csv')
Here is the corresponding traceplot:
I have the following questions:
- Every now and then, the model doesn’t start sampling because initial energy is bad. Sometimes it works, and sometimes it doesn’t. What could cause this behavior?
- The sampler clearly gets stuck. The only difference between the second model and the first is the line
TS = p_high.argmax(axis=1). If I replace it withTS = season, the models are identical and the traces look good. Why does this line create sampling problems, and is there a way to fix it? - The model uses an incredible amount of memory (>50 GB for 100 samples in 2 chains) and runs very slowly. Did I introduce a memory leak?
Any thoughts and feedback would be greatly appreciated!