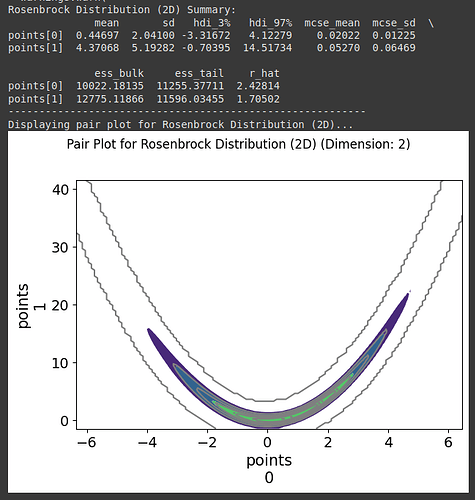
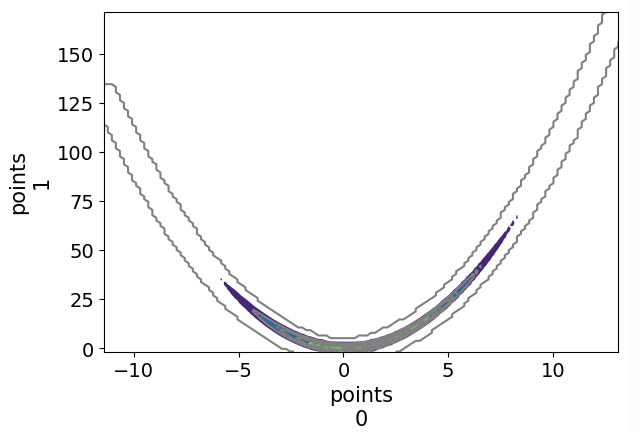
Hello !
Last weekend I watched this video about diffusion models and learned about Langevin diffusion.
(I am originally a C++ dev who is now doing a bit of python/pytorch)
This first seemed crazy to me that a random walk combined with a gradient descent on the log potential could get you this very potential (with the subtle sqrt(eps/2) mix) so I had to look into the proofs and I also had an idea about combining optimizer algorithm with Langevin diffusion.
The idea is pretty simple and after coding it up I found a paper from 2020 doing exactly what I did.
Then I learned about MALA, HMC and NUTS (this Hamiltonian idea is incredible) and I had an other idea for a sampler.
The issue with discretize langevin stepsize is you get some kind of convolution by a gaussian kernel of the posterior you are trying to sample. You can reduce the stepsize but you get much more autocorrelation.
My idea was to:
- use many // chains using the modern vectorized hardware that we have
- use adaptative gradients scheme as warmup since it would explore much faster
- use subchains with a (1-t)^2 annealing schedule after warmup keeping only the last subchain sample
edit: the schedule idea is very similar to this paper I’m always 5/6 years late…
This way you progressively deacrease the variance of the noise kernel, get your sample and start again from large variance noise kernel. Since you go from enlarge fuzzy density to a much sharper image, it looks like a “vascularisation” of the distribution, hence the codename for this sampling algorithm: Hearbeat.
I wrote the sampler in pytorch and asked Gemini to recode it in JAX (got a x3 speed improvement) and I’ve tried running the sampler on a very simple mixture of gaussian against other JAX samplers from PyMC (numpyro and blackjax), and I get very comparable result for a very different algorithm. ^^ (note I used 1024 chains for my algo and 8 for the NUTS sampler to get similar running time, 8 chains seems to get the fastest ESS/sec for the NUTS samplers)
I ran on CPU without multithread and need to do much more tests with other distributions and on GPU. From what I get, it is much easier to tune my sampler to increase num_sample/ESS where for example the acceptance probability isn’t ideal for NUTS. It is also much much faster on CPU than NUTS sampler if you need many chains, I need to check this is also the case on GPU.
It is way too early to share but I’m too excited not to so here is a notebook if you want to play with it. I will see if I can provide a better support (colab?) later this we. Any comments are welcome. ![]()
Colab link: Google Colab
Thank you for reading this far.
NB: Beware I am not a statistician and still very new to bayesian statistics.