@pymc-bot
Hello everyone (bot included!).
I am working on a problem involving constructing and inverting a matrix formed by Legendre polynomials.
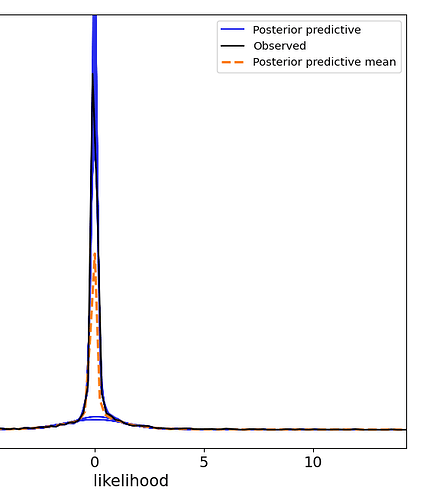
My problem is that I can see from the resulting posterior distributions, that the original values, which are generally quite close to zero, have been recovered in the peak of the distribution, however other modal peaks are observable. I would like to get rid of them.
I have experimented with Laplace and Normal distributions however the effect is the same. The run time is also quite long for this even with 1000 draws and 1000 tuning steps - suggesting to me that something inefficient is occurring.
The general thrust of the code is as follows
with pm.Model() as model:
# # Use a function to describe varying gamma
# Beta_pymc_1 = pm.Laplace("Beta_pymc_1", mu=0.1, b=0.001, shape=6)
# Beta_pymc_2 = pm.Laplace("Beta_pymc_2", mu = 0.1, b = 0.00003, shape= 6 )
# Beta_pymc_3 = pm.Laplace("Beta_pymc_3", mu =0.1, b = 0.00002 , shape= 6 )
# Beta_pymc_4 = pm.Laplace("Beta_pymc_4", mu = 0.1, b = 0.00005, shape= 6 )
Beta_pymc_1 = pm.Normal("Beta_pymc_1", mu=0.0, sigma=0.05, shape=6)
Beta_pymc_2 = pm.Normal("Beta_pymc_2", mu=0.0, sigma=0.05, shape=6)
Beta_pymc_3 = pm.Normal("Beta_pymc_3", mu=0.0, sigma=0.05, shape=6)
Beta_pymc_4 = pm.Normal("Beta_pymc_4", mu=0.0, sigma=0.05, shape=6)
# noise component
sigma_noise = pm.HalfNormal("sigma_noise", sigma= 0.5)
# Compute transformed gamma terms
G11 = pt.dot(X_pt, Beta_pymc_1)
G21 = -pt.dot(X_pt, Beta_pymc_2)
G12 = -G21
G33 = pt.dot(X_pt, Beta_pymc_3)
G43 = pt.dot(X_pt, Beta_pymc_4)
G34 = -G43
# Efficient tensor construction of Gamma_model shape (n_freq, 4, 4)
n_freq = G11.shape[0]
zero = pt.zeros_like(G11)
row0 = pt.stack([G11, G21, zero, -G43], axis=1)
row1 = pt.stack([G12, G11, zero, zero], axis=1)
row2 = pt.stack([zero, zero, G33, G34], axis=1)
row3 = pt.stack([G43, zero, -G34, G33], axis=1)
Gamma_model = pt.stack([row0, row1, row2, row3], axis=1) # shape (n_freq, 4, 4)
Gamma_model = pm.Deterministic("Gamma_model", Gamma_model)
#Next steps perform the inverse operation
# 1: obtain INV_BRACKET = pt.linalg.inv(A+ Gamma_model)
# 2: Use INV_BRACKKET within a larger matrix expression to get Y
likelihood = pm.Normal("likelihood", mu=Y, sigma = sigma_noise, observed=data)
an example of one set of Beta values to infer
0.0135227
0.00814225
0.0092286
0.00346928
-0.0111075
-0.000543785
and the post inference results.
zoomed in to the key region
If there is anyone out there who can help with this I would be most grateful.
![]()