Say we generate data by nesting a latent variable from a logistic model within a Poisson model (perhaps “nesting” is not the best word here, but I do not know a better one). For example:
import pymc3 as pm
import numpy as np
# generate data
rng = np.random.default_rng(12345678)
# Parameters of interest
a = 0.9
b = -0.7
c = -0.3
d = 0.5
x_1 = rng.random(1000)
# Latent is the logistic reg. latent variable
latent = a + x_1*b
p = 1/(1+np.exp(-latent))
# observed trials
trial_success = np.int_( rng.random(x_1.shape[0]) < p)
# Now the number of events
lam = np.exp(c + d*latent)
num_event = rng.poisson(lam=lam)
Intuitively, the way I would model this is to nest a logistic regression inside a Poisson model:
# Now both at the same time
with pm.Model() as simple_log:
a_ = pm.Normal('a', 0, sd=100)
b_ = pm.Normal('b', 0, sd=100)
c_ = pm.Normal('c', 0, sd=100)
d_ = pm.Normal('d', 0, sd=100)
# Latent specification
# Latent for the bernoulli
latent_ = pm.Deterministic(name='latent_bin', var= a_ + b_ * x_1)
# Latent for the poisson
latent_2 = pm.Deterministic(name='latent_pois', var=c_ + latent_*d_ )
# Bernoulli random vector with probability of success
pm.Bernoulli(name='logit', p=pm.invlogit(latent_), observed=trial_success)
# Poisson random vector
pm.Poisson(name='pois', mu=pm.math.exp(latent_2), observed=numbers)
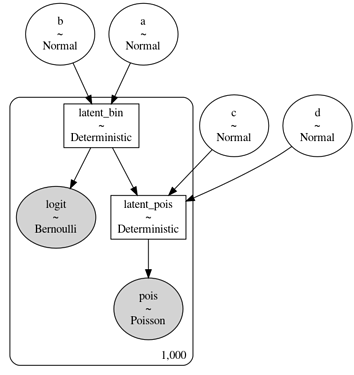
This is the DAG of the model, which corresponds to the generating process above (as far as I can understand):
However, I am having a really hard time to sample from this model, as I anticipated. Is there a good way to re-parametrize the model – or any miraculous receipt for sampling?
To stabilize sampling, I can think about ways to independently sample the logistic and Poisson part of the model. First, I can fit a standard logistic regression. Then, if I fix the a and b parameters to their posterior expected value in the Poisson part, the Poisson part becomes a standard GLM again. Yet, if I fix the a and b parameters, I am losing all information about the variability in my posteriors. So I am wondering if there is any way to incorporate that information.