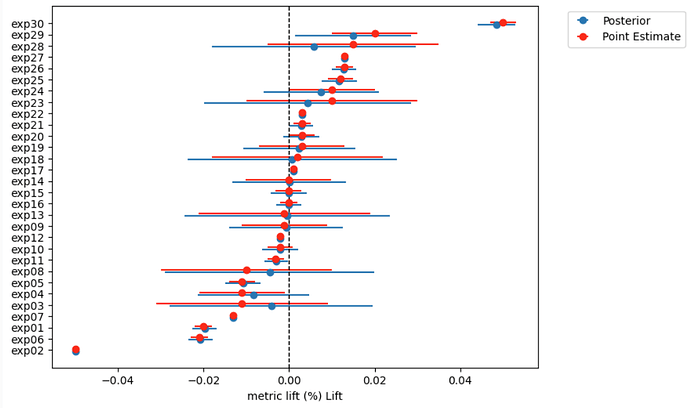
Hi all. I have the following model to pull estimated metric deltas or lifts (percentage impact) for a/b tests (experiments), with very intuitive results, where experiment lifts, which are more uncertain are pulled towards zero, while more certain experiment lifts are kept as is (see included chart)
experiment_ids = np.array([f"exp{i+1:>02.0f}"for i in range(30)])
experiment_lifts = np.array([-0.02, -0.05, -0.011, -0.011, -0.011, -0.021, -0.013, -0.01, -0.0011, -0.0021, -0.0031, -0.0021, -0.0011, -0.0001, -0.0001, -0.0001, 0.001, 0.002, 0.003, 0.003, 0.003, 0.003, 0.01, 0.01, 0.012, 0.013, 0.013, 0.015, 0.02, 0.05])
standard_errors = np.array([0.002, 0.0002, 0.02, 0.01, 0.003, 0.002, 0.0002, 0.02, 0.01, 0.003, 0.002, 0.0002, 0.02, 0.01, 0.003, 0.002, 0.0002, 0.02, 0.01, 0.003, 0.002, 0.0002, 0.02, 0.01, 0.003, 0.002, 0.0002, 0.02, 0.01, 0.003,])
with pm.Model() as normal_normal_non_centered_model:
normal_normal_non_centered_model.add_coord("experiment", experiment_ids)
lifts = pm.Data("lifts", experiment_lifts, dims="experiment")
standard_errors = pm.Data(
"standard_errors", standard_errors, dims="experiment"
)
M = pm.Normal("M", mu=0, sigma=0.02)
s = pm.HalfNormal("s", 0.02)
noise = pm.Normal(
"noise", mu=0, sigma=1, dims="experiment"
)
delta = pm.Deterministic("delta", M + s * noise, dims="experiment")
obs = pm.Normal(
"obs", mu=delta, sigma=standard_errors, observed=lifts, dims="experiment"
)
with normal_normal_non_centered_model:
idata = pm.sample(tune=15000, target_accept=0.99)
with normal_normal_non_centered_model:
idata.extend(pm.sample_posterior_predictive(idata))
posterior_pred_samples = idata.posterior_predictive["obs"]
posterior_pred_mean = posterior_pred_samples.mean(("chain", "draw"))
posterior_pred_se = posterior_pred_samples.std(axis=(0, 1))
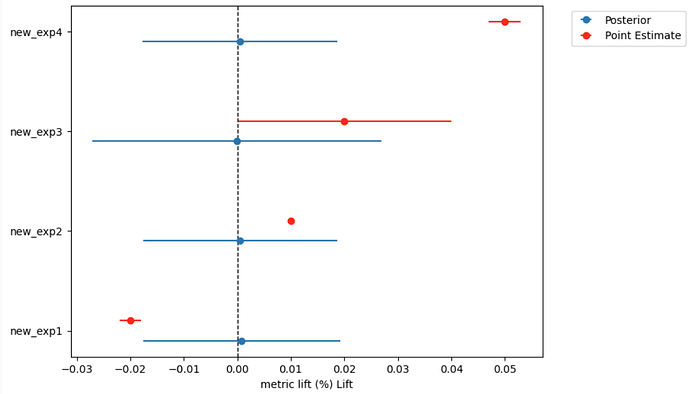
However, when I try out of sample prediction, the posterior predictions are all centered around zero and behave very differently:
new_experiment_ids = np.array(["new_exp1", "new_exp2", "new_exp3", "new_exp4"])
new_lifts = np.array([-0.02, 0.01, 0.02, 0.05])
new_standard_errors = np.array([0.002, 0.0002, 0.02, 0.003])
# Define the new coordinates
new_coords = {"experiment_variant": new_experiment_ids}
with normal_normal_non_centered_model:
# Update the dimensions to match new data
pm.set_data({"lifts": new_lifts, "standard_errors": new_standard_errors}, coords=new_coords)
# Draw posterior predictive samples using the updated model with new data
out_of_sample_predictions = pm.sample_posterior_predictive(idata, predictions=True)
posterior_pred_samples = out_of_sample_predictions.predictions["obs"]
posterior_pred_mean = posterior_pred_samples.mean(("chain", "draw"))
posterior_pred_se = posterior_pred_samples.std(axis=(0, 1))
Is it an issue with model specifications, or I am doing out of sample predictions incorrectly? Thanks a loft for you support!