Hi All,
I implemented a zero-inflated student-t distribution. Would you mind taking a look to see if it’s correct? Particularly I find the extreme values that are in both my prior- and posterior predictive checks a bit curious. I guess a student-t would allow for such values to pop up?
I would also like to know if my logp is correct. In the zero-inflated beta distributions I found here and here, when value = 0, only tt.log(self.pi) is returned. I do not understand why. Shouldn’t we also add pi * p(x=0|params) to this, like in ZeroInflatedPoisson for example?
Btw in my implementation, pi = probability of zero, so opposite to ZeroInflatedPoisson and the like.
class ZeroInflatedStudentT(pm.Continuous):
def __init__(self, nu, mu, sigma, pi, *args, **kwargs):
super(ZeroInflatedStudentT, self).__init__(*args, **kwargs)
self.nu = tt.as_tensor_variable(pm.floatX(nu))
self.mu = tt.as_tensor_variable(mu)
lam, sigma = pm.distributions.continuous.get_tau_sigma(tau=None, sigma=sigma)
self.lam = lam = tt.as_tensor_variable(lam)
self.sigma = sigma = tt.as_tensor_variable(sigma)
self.pi = pi = tt.as_tensor_variable(pm.floatX(pi))
self.studentT = pm.StudentT.dist(nu, mu, lam=lam)
def random(self, point=None, size=None):
nu, mu, lam, pi = pm.distributions.draw_values(
[self.nu, self.mu, self.lam, self.pi],
point=point, size=size)
g = pm.distributions.generate_samples(
sp.stats.t.rvs, nu, loc=mu, scale=lam**-0.5,
dist_shape=self.shape, size=size)
return g * (np.random.random(g.shape) < (1 - pi))
def logp(self, value):
logp_val = tt.switch(
tt.neq(value, 0),
tt.log1p(-self.pi) + self.studentT.logp(value),
# tt.log(self.pi))
pm.logaddexp(tt.log(self.pi), tt.log1p(-self.pi) + self.studentT.logp(0)))
return logp_val
Generate fake data:
prob_dropout = 0.3
mean_expr = 5
n = 300
dropouts = np.random.binomial(1, prob_dropout, size=n)
expr = (1 - dropouts) * sp.stats.t.rvs(df=2, loc=mean_expr, scale=3, size=n)
plt.hist(expr, bins=50);

Prior predictive
with pm.Model() as t_model:
p = pm.Normal('p', 0, 1.5)
pi = pm.Deterministic('pi', pm.math.invlogit(p))
mu = pm.Normal('mu', 0, 10)
sigma = pm.Exponential('sigma', .5)
nu = pm.Gamma('nu', alpha=5, beta=2)
obs = ZeroInflatedStudentT('obs', nu=nu, mu=mu, sigma=sigma,
pi=pi, observed=expr)
priorpred = pm.sample_prior_predictive()
fig, axs = plt.subplots(figsize=(12, 3), ncols=5)
axs[0].hist(priorpred['pi'], bins=20); axs[0].set_title('pi')
axs[1].hist(priorpred['mu'], bins=20); axs[1].set_title('mu')
axs[2].hist(priorpred['sigma'], bins=20); axs[2].set_title('sigma')
axs[3].hist(priorpred['nu'], bins=20); axs[3].set_title('nu')
axs[4].hist(priorpred['obs']); axs[4].set_title('obs') #, bins=50);
fig.tight_layout()
Sample.
with t_model:
trace = pm.sample()
pm.traceplot(trace);
Posterior predictive.
with t_model:
postpred = pm.sample_posterior_predictive(trace, var_names=['obs'])
t_idata = az.from_pymc3(trace, posterior_predictive=postpred, prior=priorpred)
az.plot_ppc(t_idata);

plt.hist(postpred['obs'].flatten(), bins=2000, alpha=.3, label='Predictive');
# Limit x-axis so we can actually see the distribution
plt.xlim(-30, 30)
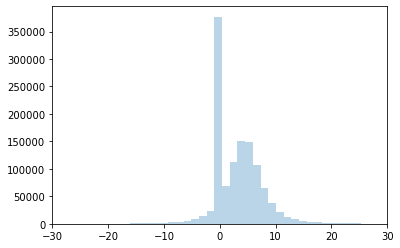
plt.hist(expr, bins=30, alpha=.3, label='Data');

Thanks!!

