I’m building a model where the group means and slopes are drawn from a mutlivariate normal distribution, with a uniform prior on the correlation between the 2.
While running prior predictive simulation, I noticed the array drawn from MVN consists of NaN and likewise all summaries associated are also NaN. I’d like to set up some sensible priors to (hopefully) speed up sampling.
Below is some code to create fake data and run the model.
import pandas as pd
import numpy as np
import pymc3 as pm
import arviz as az
N = 100
M = 10
idx = np.repeat(range(M), N)
sd_alpha = 1.5
sd_beta = 2.5
p = 0.4
cov_real = [[sd_alpha**2, sd_alpha*sd_beta*p],
[sd_alpha*sd_beta*p, sd_beta**2]]
means = [2.5, 1]
alpha_beta = np.random.multivariate_normal(means, cov = cov_real, size = M)
alpha_real = alpha_beta[:,0]
beta_real = alpha_beta[:, 1]
y_m = np.zeros(len(idx))
x_m = np.random.normal(10, 1, len(idx))
y_m = alpha_real[idx] + beta_real[idx]*x_m + eps_real
with pm.Model() as cov_m1 :
sigma = pm.HalfStudentT('sigma', nu = 3, sd = 0.5)
ab_mu = [0, 0]
sd_a = 1
sd_b = 1
p = pm.Uniform('p', -0.5, 0.5)
Cov = pm.math.stack(([sd_a**2, sd_a*sd_b*p], [sd_a*sd_b*p, sd_b**2]))
ab = pm.MvNormal('ab', mu=ab_mu, cov=Cov, shape = (10, 2))
mu = ab[:,0][idx] + ab[:,1][idx]*x_m
y_pm = pm.Normal('y_pm', mu=mu, sd=sigma, observed=y_m)
When I run the following
with cov_m1:
prior_cov_m1 = pm.sample_prior_predictive()
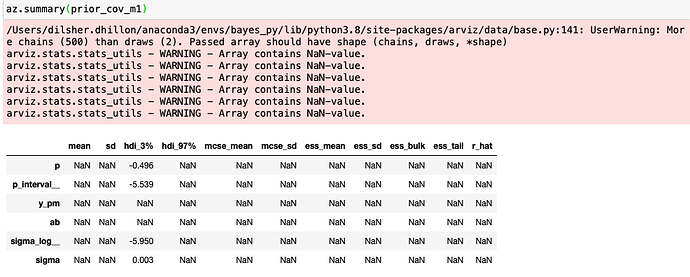
az.summary(prior_cov_m1)
I get the following.
Similarly
Appreciate any insight into what I’m missing in the model statement in order to simulate from the prior. I’ve tried even tighter priors but the problem persists.
Thank you