Thank you for your response! My most recent problem is a two connected hierarchical models, small dataset here:
School,StudentID,RepeatID,science_test1,science_test2
Jefferson,A,First Try,98.63,96.24
Jefferson,A,Second Try,99.35,109.69
Jefferson,A,Third Try,102.06,94.72
Jefferson,B,First Try,102.75,102.53
Jefferson,B,Second Try,104.93,97.91
Jefferson,B,Third Try,92.75,99.62
Jefferson,C,First Try,108.93,131.58
Jefferson,C,Second Try,98.98,88.41
Jefferson,C,Third Try,92.75,85.96
Jefferson,D,First Try,109.99,120.98
Jefferson,D,Second Try,91.21,85.17
Jefferson,D,Third Try,99.68,97.05
Washington,E,First Try,9.05,3.7
Washington,E,Second Try,0.46,0.85
Washington,E,Third Try,0.68,1.69
Washington,F,First Try,0.15,0.2
Washington,F,Second Try,0.75,0.8
Washington,F,Third Try,0.43,0.46
Washington,G,First Try,1.24,1.35
Washington,G,Second Try,0.7,1.11
Washington,G,Third Try,1.58,1.49
Washington,H,First Try,7.5,0.5
Washington,H,Second Try,0.59,0.49
Washington,H,Third Try,0.43,0.47
I want to test if there is a difference in scientific ability between the Jefferson school and the Washington school as measured by both of the science tests (NOTE: in the full dataset there are more than 2 schools). In each of the 2 schools each of 4 students takes each of 2 tests 3 different times. I am able to estimate one beta for each school-test pair and pool into two betas one level above the test level:

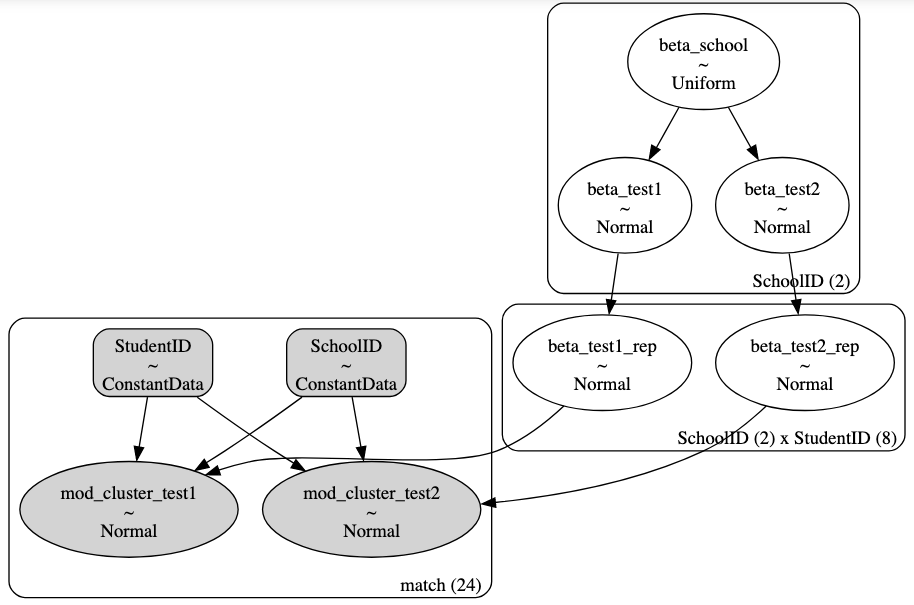
But this model doesn’t cluster scores at the StudentID level and that clustering is what I am struggling with. I want to estimate a model like this:
I think I’m encoding the hierarchy correctly but something is going wrong with how I’m giving PyMC the indicators for SchoolID and StudentID and I don’t understand why. When I try to sample this model it gives a ValueError: Incompatible Elemwise input shapes.
How does this clustering work with PyMC? In the final model call I’m telling PyMC to cluster by school and student by subsetting a pm.Normal object like:
mod_test1 = pm.Normal("mod_cluster_test1",
mu = beta_test1_rep[school, student],
sigma = 2,
observed = df.science_test1,
dims = "match")
But I suspect this is incorrect, or at least not flexible enough. It’s only this one model that’s going wrong because I’ve done this double subsetting before and it’s worked just fine. Perhaps I’ve encoded something else wrong in this model?