Dear there,
I am trying to use Bayesian hierarchical regression models (or Bayesian linear mixed model) to analyze a small dataset that we usually do with repeated-measure ANOVAs. I could have run it using BRMs, but I decide to use PyMC3 because I think PyMC3 will force me to learn the details about constructing a model. However, I am new to Bayesian hierarchical/multi-level models, so I am trying my best to describe my question. If you feel that my description is not clear enough, please point it out, and I will add more info.
My data are from 25 participants’ subjective ratings in a 2 by 2 within-subject design (i.e., 4 conditions in total). Each participant has 4 ratings for each condition (i.e., 16 ratings in total). The ratings are from 0 ~ 99, with many zeros for some conditions.
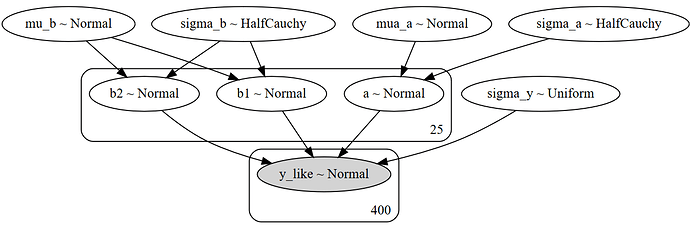
I first tried to follow this tutorial: https://docs.pymc.io/notebooks/multilevel_modeling.html. It worked well until I tried to include the interaction between two variables. So the last model I constructed is like this:
with Model() as example_LMM:
# priors
mu_a = Normal('mua_a', mu=0., sigma=1e5)
sigma_a = HalfCauchy('sigma_a', 5)
mu_b = Normal('mu_b', mu=0., sigma=1e5)
sigma_b = HalfCauchy('sigma_b', 5)
# Random intecepts
a = Normal('a', mu=mu_a, sigma=sigma_a, shape=subjs)
b1 = Normal('b1', mu=mu_b, sigma=sigma_b, shape=subjs)
b2 = Normal('b2', mu=mu_b, sigma=sigma_b, shape=subjs)
# Model error
sigma_y = Uniform('sigma_y', lower = 0, upper=100)
# expected value
y_hat = a[subj] + b1[subj]*blockType + b2[subj]*CSType
# Data likelihood
y_like = Normal('y_like', mu=y_hat, sigma=sigma_y, observed=AnxRatings)
model_to_graphviz(example_LMM)
Then I read these two posts
And I tried to imitate the model from the first one (thanks @Jack_Caster for sharing the notebook), and wrote a model like this:
(mx_en, mx_ex) = pt.dmatrices("AnxRating ~ blockType * CSType", data=df_d1, return_type='dataframe')
with Model() as mdl_interaction:
# define priors weakly informative Normal
# Fixed effects
#mu_a = Normal('mua_a', mu=0., sigma=100)
#sigma_a = HalfCauchy('sigma_a', 5)
#mu_b = Normal('mu_b', mu=0., sigma=100)
#sigma_b = HalfCauchy('sigma_b', 5)
beta_X_intercept = pm.HalfNormal('beta_X_intercept', sd=1000) # contrain it to positive values
beta1_X_slope = pm.Normal('beta1_X_slope', mu=0, sd=100)
beta2_X_slope = pm.Normal('beta2_X_slope', mu=0, sd=100)
beta3_X_slope = pm.Normal('beta3_X_slope', mu=0, sd=100)
beta_X = tt.stack(beta_X_intercept, beta1_X_slope, beta2_X_slope, beta3_X_slope)
estimate_X = pm.math.dot(X, beta_X)
# Random intecepts
#a = Normal('a', mu=mu_a, sigma=sigma_a, shape=subjs)
#b1 = Normal('b1', mu=mu_b, sigma=sigma_b, shape=subjs)
#b2 = Normal('b2', mu=mu_b, sigma=sigma_b, shape=subjs)
#b3 = Normal('b3', mu=mu_b, sigma=sigma_b, shape=subjs)
## Random effect
sigma_Z = pm.HalfNormal('sigma_Z', sd=100)
gamma_Z_raw = pm.Normal('gamma_Z_raw', mu=0, sd=1, shape=Z.shape[1])
gamma_Z = pm.Deterministic('gamma_Z', gamma_Z_raw * sigma_Z)
estimate_Z = pm.math.dot(Z, gamma_Z)
# Model error
#sigma_y = Normal('sigma_y', mu = 0, sigma=1e6)
# expected value
#y_hat = (a[subj] +
# b1[subj]*mx_ex['blockType'] +
# b2[subj]*mx_ex['CSType'] +
# b3[subj]*mx_ex['blockType:CSType'])
mu_estimate = pm.Deterministic('mu_estimate', estimate_X + estimate_Z)
sigma_unexplained = pm.HalfNormal('sigma_unexplained', sd=1000) # unexplained variability
y_likelihood = pm.Normal('y_likelihood', mu=mu_estimate, sd=sigma_unexplained, observed=AnxRatings)
# Data likelihood
#y_like = Normal('y_like', mu=y_hat, sigma=sigma_y, observed=AnxRatings)
model_to_graphviz(mdl_interaction)
I also realized that the model from https://docs.pymc.io/notebooks/multilevel_modeling.html (let’s call this as example 1) and Hierarchical Linear Regression Model -- why is it not converging? (let’s call this as example 2) are different.
I felt that there are a few points that I am not very clear so that I need some guide, maybe just pointing to me where can I found the answer. These points are:
(1) If I understand correctly, example 2 includes both the fixed effect & random effect, example 1 only includes the random effect? I am a bit confusing: are we supposed to always include both fixed and random effect?
(2) For example 2 (Hierarchical Linear Regression Model -- why is it not converging?) and here (Constructing the random effects model matrix Z), the deterministic function were included, when should we use this?
(3) How to plot the traces of random effect together. I as saw @Jack_Caster’s note book (https://github.com/JackCaster/GLM_with_PyMC3/blob/master/notebooks/LME%20-%20Sleep%20study.ipynb), the trace of random effect gamma_Z were plotted together:
But when I tried to plot this notebook in my own PC, seems they are in different subplots.
(4) I knew that Normal distribution is not appropriate for my data (ratings), what would be a better choice?
Many thanks.