Hello,
I have followed an article by Chad Scherrer on Bayesian Optimal Pricing and managed to get it to work well with my own data. I’ve also bound the beta variable to be a negative number. Using sales data for a single item, the model works as expected and the posterior prediction matches closely with the real data.
When I change the code going from examples provided in this forum on how to change it to a multivariate model, the values for beta match closely with the single item model, but the alpha is completely different, and if I feed these values into a formula to predict the quantity given a price, I get back values that are much larger. I am wanting to create a independent model so I can quickly get the elasticities for multiple items at the same time.
Single item model:
item_data = [{'p': 75.1, 'q': 54}, {'p': 78.64, 'q': 52}, {'p': 90.39, 'q': 21}, {'p': 96.06, 'q': 15}, {'p': 91.48, 'q': 18}, {'p': 89.76, 'q': 23}, {'p': 95.17, 'q': 15}, {'p': 86.36, 'q': 38}, {'p': 90.08, 'q': 25}, {'p': 93.89, 'q': 15}, {'p': 81.67, 'q': 44}, {'p': 83.6, 'q': 41}, {'p': 81.12, 'q': 33}, {'p': 82.81, 'q': 35}, {'p': 83.17, 'q': 29}, {'p': 88.87, 'q': 10}, {'p': 84.73, 'q': 16}, {'p': 82.1, 'q': 14}, {'p': 86.86, 'q': 20}, {'p': 94.4, 'q': 17}, {'p': 97.45, 'q': 24}, {'p': 89.41, 'q': 38}, {'p': 79.23, 'q': 132}, {'p': 73.53, 'q': 113}, {'p': 86.4, 'q': 44}, {'p': 78.98, 'q': 39}, {'p': 62.91, 'q': 78}, {'p': 62.92, 'q': 89}, {'p': 72.88, 'q': 130}, {'p': 71.8, 'q': 81}, {'p': 75.1, 'q': 53}, {'p': 75.75, 'q': 61}, {'p': 78.71, 'q': 35}, {'p': 78.94, 'q': 35}, {'p': 81.05, 'q': 55}, {'p': 64.71, 'q': 407}, {'p': 56.07, 'q': 277}, {'p': 64.14, 'q': 130}, {'p': 78.84, 'q': 82}, {'p': 82.47, 'q': 114}, {'p': 76.08, 'q': 72}, {'p': 86.98, 'q': 30}, {'p': 86.39, 'q': 37}, {'p': 82.04, 'q': 36}, {'p': 80.49, 'q': 36}, {'p': 82.78, 'q': 36}, {'p': 73.1, 'q': 42}, {'p': 76.94, 'q': 47}, {'p': 71.04, 'q': 59}, {'p': 80.27, 'q': 48}, {'p': 84.19, 'q': 40}]
items = pd.DataFrame(item_data)
with pm.Model() as m:
a = pm.Cauchy('a',0,5)
NegCauchy = pm.Bound(pm.Cauchy, lower=-np.inf, upper=0)
b = NegCauchy('b',0,.5)
logμ0 = a + b * (np.log(items.p) - np.log(items.p).mean())
μ0 = pm.Deterministic('μ0',np.exp(logμ0))
qval = pm.Poisson('q0',μ0,observed=items.q)
with m:
t = pm.sample(1000, cores=1, tune=750)
plt.plot(t['a'],t['b'],'.',alpha=0.5);
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (2 chains in 1 job)
NUTS: [b, a]
Sampling chain 0, 0 divergences: 100%|███████████████████████████████████████████| 1750/1750 [00:00<00:00, 1978.44it/s]
Sampling chain 1, 0 divergences: 100%|███████████████████████████████████████████| 1750/1750 [00:00<00:00, 2179.24it/s]

p = np.linspace(items.p.min() * .8, items.p.max() * 1.2, num=100)
np.exp(t.a + t.b * (np.log(p).reshape(-1,1) - np.log(items.p).mean()))
array([[ 965.68164247, 841.48493256, 1044.10041234, ..., 1023.8077616 ,
1022.91339029, 1003.82476968],
[ 889.34484971, 778.10209929, 959.9691476 , ..., 941.25641286,
940.21155391, 922.88566178],
[ 820.11200721, 720.38687373, 883.79282242, ..., 866.51496749,
865.35144519, 849.60385152],
...,
[ 7.66359729, 8.46561696, 7.51595867, ..., 7.34453369,
7.23680109, 7.20162925],
[ 7.4211787 , 8.21076235, 7.273494 , ..., 7.10743644,
7.00253458, 6.96914814],
[ 7.18787135, 7.9651006 , 7.04029305, ..., 6.87940304,
6.77724447, 6.74555433]])
Same item in a multivariate independent model:
items_data = [{'item': 0, 'p': 75.1, 'q': 54}, {'item': 1, 'p': 4.81, 'q': 38}, {'item': 0, 'p': 78.64, 'q': 52}, {'item': 1, 'p': 4.23, 'q': 39}, {'item': 0, 'p': 90.39, 'q': 21}, {'item': 1, 'p': 4.74, 'q': 26}, {'item': 0, 'p': 96.06, 'q': 15}, {'item': 1, 'p': 4.76, 'q': 33}, {'item': 0, 'p': 91.48, 'q': 18}, {'item': 1, 'p': 4.83, 'q': 44}, {'item': 0, 'p': 89.76, 'q': 23}, {'item': 1, 'p': 4.79, 'q': 56}, {'item': 0, 'p': 95.17, 'q': 15}, {'item': 1, 'p': 4.7, 'q': 47}, {'item': 0, 'p': 86.36, 'q': 38}, {'item': 1, 'p': 4.7, 'q': 57}, {'item': 0, 'p': 90.08, 'q': 25}, {'item': 1, 'p': 4.83, 'q': 65}, {'item': 0, 'p': 93.89, 'q': 15}, {'item': 1, 'p': 4.77, 'q': 71}, {'item': 0, 'p': 81.67, 'q': 44}, {'item': 1, 'p': 4.75, 'q': 66}, {'item': 0, 'p': 83.6, 'q': 41}, {'item': 1, 'p': 4.85, 'q': 65}, {'item': 0, 'p': 81.12, 'q': 33}, {'item': 1, 'p': 4.72, 'q': 66}, {'item': 0, 'p': 82.81, 'q': 35}, {'item': 1, 'p': 4.82, 'q': 68}, {'item': 0, 'p': 83.17, 'q': 29}, {'item': 1, 'p': 4.61, 'q': 75}, {'item': 0, 'p': 88.87, 'q': 10}, {'item': 1, 'p': 4.54, 'q': 64}, {'item': 0, 'p': 84.73, 'q': 16}, {'item': 1, 'p': 4.59, 'q': 86}, {'item': 0, 'p': 82.1, 'q': 14}, {'item': 1, 'p': 4.16, 'q': 87}, {'item': 0, 'p': 86.86, 'q': 20}, {'item': 1, 'p': 4.61, 'q': 72}, {'item': 0, 'p': 94.4, 'q': 17}, {'item': 1, 'p': 4.81, 'q': 67}, {'item': 0, 'p': 97.45, 'q': 24}, {'item': 1, 'p': 4.6, 'q': 53}, {'item': 0, 'p': 89.41, 'q': 38}, {'item': 1, 'p': 4.59, 'q': 60}, {'item': 0, 'p': 79.23, 'q': 132}, {'item': 1, 'p': 4.76, 'q': 58}, {'item': 0, 'p': 73.53, 'q': 113}, {'item': 1, 'p': 4.16, 'q': 55}, {'item': 0, 'p': 86.4, 'q': 44}, {'item': 1, 'p': 4.17, 'q': 60}, {'item': 0, 'p': 78.98, 'q': 39}, {'item': 1, 'p': 4.08, 'q': 44}, {'item': 0, 'p': 62.91, 'q': 78}, {'item': 1, 'p': 3.12, 'q': 49}, {'item': 0, 'p': 62.92, 'q': 89}, {'item': 1, 'p': 3.11, 'q': 68}, {'item': 0, 'p': 72.88, 'q': 130}, {'item': 1, 'p': 4.24, 'q': 61}, {'item': 0, 'p': 71.8, 'q': 81}, {'item': 1, 'p': 4.38, 'q': 41}, {'item': 0, 'p': 75.1, 'q': 53}, {'item': 1, 'p': 4.02, 'q': 57}, {'item': 0, 'p': 75.75, 'q': 61}, {'item': 1, 'p': 4.29, 'q': 49}, {'item': 0, 'p': 78.71, 'q': 35}, {'item': 1, 'p': 4.31, 'q': 58}, {'item': 0, 'p': 78.94, 'q': 35}, {'item': 1, 'p': 4.35, 'q': 41}, {'item': 0, 'p': 81.05, 'q': 55}, {'item': 1, 'p': 4.29, 'q': 47}, {'item': 0, 'p': 64.71, 'q': 407}, {'item': 1, 'p': 4.18, 'q': 69}, {'item': 0, 'p': 56.07, 'q': 277}, {'item': 1, 'p': 4.35, 'q': 54}, {'item': 0, 'p': 64.14, 'q': 130}, {'item': 1, 'p': 4.25, 'q': 54}, {'item': 0, 'p': 78.84, 'q': 82}, {'item': 1, 'p': 3.55, 'q': 65}, {'item': 0, 'p': 82.47, 'q': 114}, {'item': 1, 'p': 3.74, 'q': 76}, {'item': 0, 'p': 76.08, 'q': 72}, {'item': 1, 'p': 3.8, 'q': 67}, {'item': 0, 'p': 86.98, 'q': 30}, {'item': 1, 'p': 3.2, 'q': 57}, {'item': 0, 'p': 86.39, 'q': 37}, {'item': 1, 'p': 3.13, 'q': 67}, {'item': 0, 'p': 82.04, 'q': 36}, {'item': 1, 'p': 4.23, 'q': 53}, {'item': 0, 'p': 80.49, 'q': 36}, {'item': 1, 'p': 4.4, 'q': 49}, {'item': 0, 'p': 82.78, 'q': 36}, {'item': 1, 'p': 4.29, 'q': 36}, {'item': 0, 'p': 73.1, 'q': 42}, {'item': 1, 'p': 3.91, 'q': 56}, {'item': 0, 'p': 76.94, 'q': 47}, {'item': 1, 'p': 4.19, 'q': 49}, {'item': 0, 'p': 71.04, 'q': 59}, {'item': 1, 'p': 3.59, 'q': 58}, {'item': 0, 'p': 80.27, 'q': 48}, {'item': 1, 'p': 3.22, 'q': 78}, {'item': 0, 'p': 84.19, 'q': 40}, {'item': 1, 'p': 3.13, 'q': 56}]
items = pd.DataFrame(items_data)
n_items = items.item.nunique()
with pm.Model() as m:
a = pm.Cauchy('a',0,5,shape=n_items)
NegCauchy = pm.Bound(pm.Cauchy, lower=-np.inf, upper=0)
b = NegCauchy('b',0,.5,shape=n_items)
logμ0 = a[items.item] + b[items.item] * (np.log(items.p) - np.log(items.p).mean())
μ0 = pm.Deterministic('μ0', np.exp(logμ0))
qval = pm.Poisson('q0', μ0, observed=items.q)
with m:
t = pm.sample(1000, cores=1, tune=750)
plt.plot(t['a'][:,0],t['b'][:,0],'.',alpha=0.5)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (2 chains in 1 job)
NUTS: [b, a]
Sampling chain 0, 41 divergences: 100%|███████████████████████████████████████████| 1750/1750 [00:08<00:00, 207.04it/s]
Sampling chain 1, 3 divergences: 100%|████████████████████████████████████████████| 1750/1750 [00:10<00:00, 165.94it/s]
There were 41 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.6383162669604235, but should be close to 0.8. Try to increase the number of tuning steps.
There were 44 divergences after tuning. Increase `target_accept` or reparameterize.
The estimated number of effective samples is smaller than 200 for some parameters.
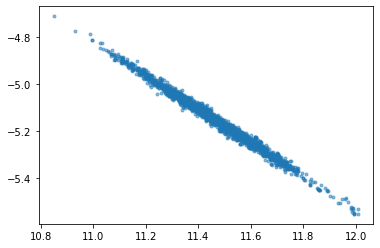
t.a[:, 0]
array([11.67, 11.65, 11.25, ..., 11.20, 11.33, 11.49])
t.b[:, 0]
array([-5.30, -5.28, -5.01, ..., -4.97, -5.07, -5.17])
pi = items.loc[items.item == 0].p
p = np.linspace(pi.min() * .9, pi.max() * 1.1, num=100)
np.exp(t.a[:,0] + t.b[:,0] * (np.log(p).reshape(-1,1) - np.log(pi).mean()))
array([[1380304.13, 1346367.87, 795723.19, ..., 742055.83, 881985.02,
1085577.98],
[1300175.60, 1268386.24, 751982.49, ..., 701535.30, 832900.29,
1023999.46],
[1225516.74, 1195717.65, 711094.97, ..., 663643.39, 787050.30,
966543.95],
...,
[27024.56, 26603.00, 19312.53, ..., 18471.94, 20412.33, 23316.39],
[26264.13, 25856.15, 18798.38, ..., 17983.48, 19862.05, 22675.53],
[25529.00, 25134.11, 18300.57, ..., 17510.45, 19329.44, 22055.58]])