Hi all, I am new to pymc and this is last step of my masters’s thesis. So whether this code worked is reallly important to me.
I am trying to fit parameters x in this form:

coefficients a is given as form of array.
I have a observed data like:
data = np.array([0.00882,0.02679, 0.04765, 0.07059, 0.09581,
0.11891, 0.14474, 0.17651, 0.19992, 0.22293, 0.2525, 0.27134,
0.29675, 0.32454, 0.34271, 0.36829, 0.38142, 0.40989])
every point in data has a linear regression mentioned above(different array a).
which means I need to find a set of parameters x to fit different points in data together best.
Now i = 11 I will set 11 priors, suppose they are all Normal~(0, 0.05)
my code is:
with pm.Model() as model:
theta = pm.Normal('theta',mu=0,sigma=0.1,shape=11)
x=theta
smiu = []
for nm in range(len(A)):
A1 = A[nm]
sum1 = 0
sum2 = 0
sum3 =0
former = 0
for i in range(11):
sum1 = x[i] * A1[i] + sum1 #
d = 11
for i in range(11):
for j in range(i, 11):
sum2 = A1[d] * x[i] * x[j] + sum2
d = d + 1
sum3 = math.log(curr_initial[nm]*1000) + sum1 + sum2 # *math.log(abs(curr_in))
smiu.append(sum3)
like = pm.Normal('like',mu=smiu, sigma=0.05,observed=data)
trace = pm.sample(draws=2000,nuts_sampler="numpyro", progressbar=False,tune=1000)
pm.sample_posterior_predictive(trace, extend_inferencedata=True)
trace.posterior_predictive
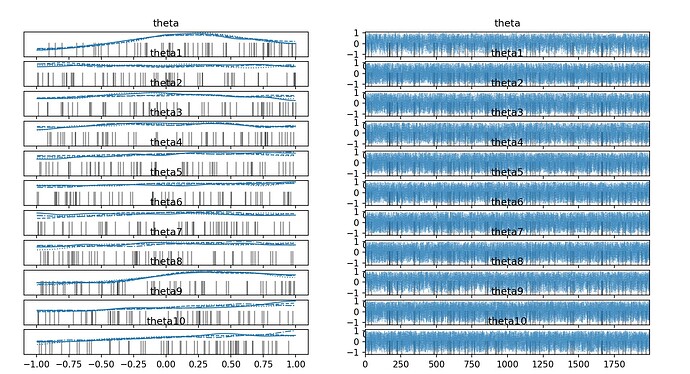
az.plot_ppc(trace, num_pp_samples=1000)
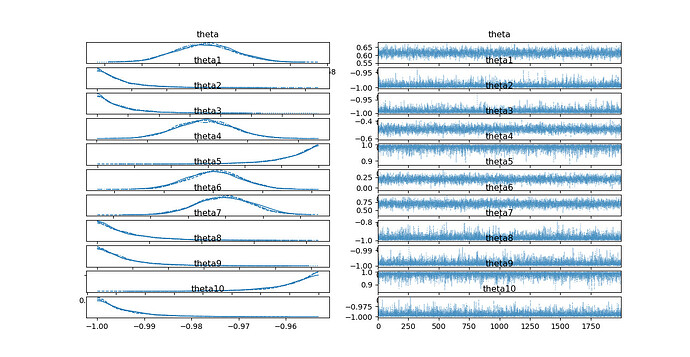
az.plot_trace(trace)
print(pm.summary(trace)[['mean', 'sd', 'mcse_mean', 'mcse_sd', 'r_hat']], 'trace')
az.plot_posterior(trace)
plt.show()
results are:
the result shows the standard deviation of posterior distribution nearly remains the same compared to prior. Is anything I did wrong? Is there any way to small the standard deviation?
I need the posterior distribution to be tall and thin.
I am using pymc5 python3.10 in windows.
the MWE is:
RS_bayesian.py (30.8 KB)
Thanks for any advice. ![]()